special tutorial
Written by: Valentin Christiaens.
Last update: 2022/08/04
IMPORTANT: to run this jupyter notebook tutorial, you will have to download the special-extras repository, and keep the utils.py file (containing utility routines specific to the dataset used in the notebook) in the same folder as the notebook. Alternatively, you can execute this notebook on Binder (in the tutorials directory).
Table of contents
Let’s start by importing a couple of useful packages. It is necessary to have both the utils.py file located in the same directory as this notebook, and the datasets in a folder at the same level as the parent directory to this notebook. This is the default if you download the GitHub repository and run the notebook from within your local copy of the repository, or if you use Binder.
[1]:
import special
special.__version__
[1]:
'0.1.5'
[2]:
%matplotlib inline
from matplotlib import pyplot as plt
import numpy as np
from pathlib import Path
1. Loading the data
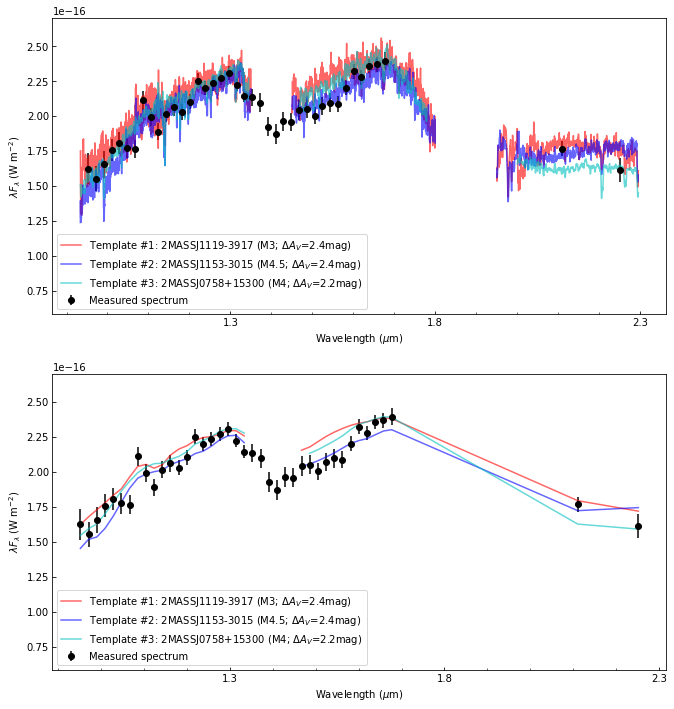
The data used in this tutorial correspond to (i) the 2MASS J19005804-3645048 b/B (aka. CrA-9 b/B) spectrum acquired from VLT/SPHERE-IFS, VLT/SPHERE-IRDIS and VLT/NACO high-contrast imaging data, and (ii) the SPHERE-IFS PSF-subtracted datacube, presented in Christiaens et al. (2021). You can find them in the ‘dataset’ folder of the special_extras repository:
CrA-9b_spectrum.fits: the measured spectrum of (sub)stellar companion CrA-9 b/B, as derived using the negative fake companion technique (Wertz et al. 2017; i.e. finding the negative flux that minimizes the absolute residuals at the location of the companion) in each IFS spectral channel and in each broad-band filter image obtained with IRDIS and NACO;CrA-9_2019_IFS_pca_annulus.fits: the post-processed Integral Field Spectrograph (IFS) datacube on CrA-9, where the stellar halo and speckles have been modeled by principal component analysis (PCA; Amara & Quanz 2012) and removed in each of the 39 spectral channels.
Let’s define our input path:
[3]:
par_path = Path('__file__').parent
rel_path = '../datasets/'
datpath = str((par_path / rel_path).resolve())+'/'
In the post-processed IFS cube, the emission from the central star has been modeled and subtracted using the ADI strategy in each spectral channel. The post-processing was performed using principal component analysis (PCA) applied in a single annulus. The annulus was chosen to encompass the faint (sub)stellar companion CrA-9b/B, which can be seen towards the left of the images.
Let’s first inspect the IFS cube using the info_fits utility implemented in the fits module:
[4]:
from special.fits import info_fits
info_fits(datpath+'CrA-9_2019_IFS_pca_annulus.fits')
Filename: /Users/valentin/GitHub/special_extras/datasets/CrA-9_2019_IFS_pca_annulus.fits
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 7 (291, 291, 39) float64
The fits file contains an IFS datacube of 39 images with wavelengths spanning 0.95 to 1.68 \(\mu\)m. Each image is 291x291 px across. Let’s now open it and check a few spectral channels.
[5]:
from special.fits import open_fits
ifs_cube = open_fits(datpath+'CrA-9_2019_IFS_pca_annulus.fits')
Fits HDU-0 data successfully loaded. Data shape: (39, 291, 291)
[6]:
from special.config import figsize
fig, axes = plt.subplots(1,3,figsize=figsize)
axes[0].imshow(ifs_cube[1])
axes[1].imshow(ifs_cube[19])
axes[2].imshow(ifs_cube[-1])
[6]:
<matplotlib.image.AxesImage at 0x7fd0d0a9d850>
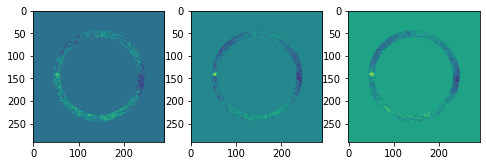
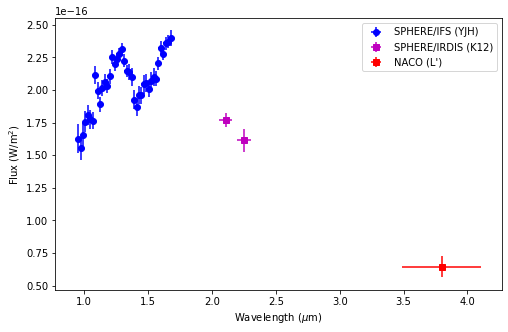
A point source can be seen towards the left of the image. This is CrA-9 b/B, a faint companion to the pre-main sequence star CrA-9, whose nature is still debated (Christiaens et al. 2021). In addition to this IFS datacube obtained with VLT instrument SPHERE/IFS, three additional detections of the companion were obtained with VLT imagers SPHERE/IRDIS at 2.11 \(\mu\)m and 2.25\(\mu\)m, and NACO at 3.8 \(\mu\)m. The full measured spectrum contains thus 42 points:
[7]:
info_fits(datpath+'CrA-9b_spectrum.fits')
Filename: /Users/valentin/GitHub/special_extras/datasets/CrA-9b_spectrum.fits
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 6 (42, 4) float32
The 4 dimensions correspond to:
the wavelength of each measurement (in \(\mu\)m);
the channel width (or FWHM of the broad band filter used in the case or IRDIS and NACO);
the flux of the companion;
the error on the flux.
[8]:
spectrum_b = open_fits(datpath+'CrA-9b_spectrum.fits')
lbda = spectrum_b[0]
dlbda = spectrum_b[1]
spec = spectrum_b[2]
spec_err = spectrum_b[3]
Fits HDU-0 data successfully loaded. Data shape: (4, 42)
Let’s visualize the measured spectrum in \(\lambda F_{\lambda}\) units:
[9]:
fig = plt.figure(figsize=figsize)
plt.errorbar(lbda[:-3], lbda[:-3]*spec[:-3], lbda[:-3]*spec_err[:-3], dlbda[:-3], 'bo',
label = 'SPHERE/IFS (YJH)')
plt.errorbar(lbda[-3:-1], lbda[-3:-1]*spec[-3:-1], lbda[-3:-1]*spec_err[-3:-1], dlbda[-3:-1], 'ms',
label = 'SPHERE/IRDIS (K12)')
plt.errorbar(lbda[-1], lbda[-1]*spec[-1], lbda[-1]*spec_err[-1], dlbda[-1], 'rs',
label = "NACO (L')")
plt.xlabel(r"Wavelength ($\mu$m)")
plt.ylabel(r"Flux (W/m$^2$)")
plt.legend()
[9]:
<matplotlib.legend.Legend at 0x7fd0d0d2c7c0>
2. Spectral correlation matrix
2.1 Estimating the spectral correlation between IFS channels
Before fitting different models to the spectrum, let’s first estimate the spectral correlation between the IFS channels. By design of the IFS, the flux at a given wavelength leaks on/overlaps with neighbouring channels. These can therefore not be considered as independent measurements. The spectral correlation matrix has therefore to be considered when fitting a given IFS spectrum. Not taking it into account can indeed lead to significant biases, and in particular to erroneous results in terms of best-fit model parameters (see e.g. Greco & Brandt 2016).
Estimating the spectral correlation can be done easily in special using the spectral_correlation routine, which follows the method recommended in Greco & Brandt (2016). For that let’s first determine the location of the companion in the IFS images, by zooming on the median image of the cube:
[10]:
x_i = 35
y_i = 130
sz = 31
plt.imshow(np.median(ifs_cube,axis=0)[y_i:y_i+sz,x_i:x_i+sz])
plt.gca().invert_yaxis()
plt.grid()
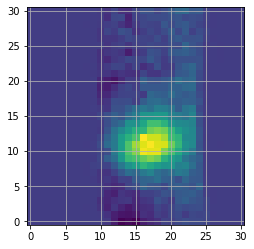
Based on the above inset, we can set the approximate coordinates of the companion, and infer its radial separation:
[11]:
pl_xy = ((x_i+17,y_i+11),) # set pl_xy as a tuple of 2-element tuples - each tuple corresponds to a different companion
cy = (ifs_cube.shape[1]/2.)-0.5
cx = (ifs_cube.shape[2]/2.)-0.5
r0 = np.sqrt((pl_xy[0][0]-cx)**2+(pl_xy[0][1]-cy)**2)
r0
[11]:
93.08598175880189
The spectral correlation depends on the radial separation to the central star. The algorithm implemented in special can therefore compute it in concentric annuli between given inner and outer radii (by default spanning the whole field). Here since we are only interested in the spectral correlation at the radial separation of the companion, let’s set the annular width, inner and outer radii such that they only encompass the companion radial separation:
[12]:
awidth=2
r_in = int(r0-awidth/2)
r_out = int(r0+awidth/2)
Furthermore, we mask the area around the companion to not bias the spectral correlation estimate with companion flux. We set the mask radius to be 4 times the FWHM, with a FWHM set to 4px:
[13]:
mask_r=4
fwhm=4
Now, we can estimate the spectral correlation at the radial separation of CrA-9b/B:
[14]:
from special.spec_corr import spectral_correlation
sp_corr = spectral_correlation(ifs_cube, awidth=awidth, r_in=r_in, r_out=r_out, pl_xy=pl_xy,
mask_r=mask_r, fwhm=fwhm)
sp_corr = sp_corr[0]
Let’s visualize it:
[15]:
plt.imshow(sp_corr)
#plt.savefig("spectral_corr_matrix.pdf", bbox_inches='tight')
plt.colorbar()
plt.gca().invert_yaxis()
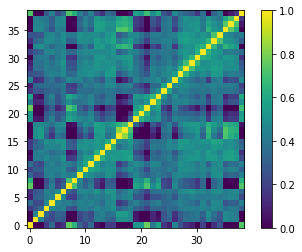
Here we used a high number of principal components for PCA, and the companion is far away from the star, such that the correlated speckle noise is efficiently subtracted from each reduced channel. Consequently, not much correlation can be seen between channels. Each spectral channel is most correlated to itself, and at most to its 2 closest neighbouring channels. Note that zero is adopted for uncorrelated channels.
2.2 Concatenating spectral correlation matrices
Since our full spectrum of CrA-9b/B also contains photometric points measured by SPHERE/IRDIS (2) and NACO (1), we need to pad the IFS spectral correlation matrix with 3 additional rows and columns of zeroes + ones on the diagonal, in order to get the final spectral correlation matrix.
This can easily be done with the combine_spec_corrs routine:
[16]:
from special.spec_corr import combine_spec_corrs
n_ird=2
n_naco=1
arr_list = [sp_corr]+[1]*n_ird+[1]*n_naco
final_sp_corr = combine_spec_corrs(arr_list)
Let’s visualize the final matrix:
[17]:
plt.imshow(final_sp_corr)
plt.colorbar()
plt.gca().invert_yaxis()
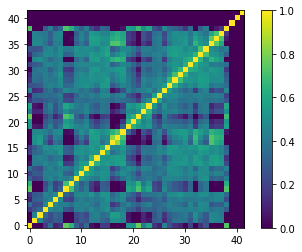
Uncomment the lines in the following box if you wish to save the final spectral correlation matrix in fits format.
[18]:
#from special.fits import write_fits
#write_fits(datpath+"CrA-9_sp_corr_matrix.fits", final_sp_corr)
3. Preliminary spectral analysis
A number of utilities are implemented in the utils_spec and spec_indices module, which allow for a preliminary qualitative analysis of the spectrum of the companion. However, to avoid biased conclusions due to extinction in this preliminary analysis, let’s first deredden the observed spectrum using the value quoted in the literature for CrA-9 (\(A_V = 1.6\) mag; Dunham et al. 2015).
3.1 Dereddening
Provided a given \(A_V\) extinction value, the extinction routine can estimate the corresponding extinction factor at different wavelengths following the Cardelli et al. (1989) law, and for a desired \(R_V\) value. Let’s consider the standard ISM value of \(R_V = 3.1\) mag (default if not provided).
[19]:
from special.utils_spec import extinction
AV_est = 1.6 #mag
A_lambda = extinction(lbda, AV_est, RV=3.1)
extinc_fac = np.power(10,-A_lambda/2.5)
Let’s plot the extinction factor as a function of wavelength:
[20]:
plt.plot(lbda, extinc_fac)
plt.xlabel(r"Wavelength ($\mu$m)")
plt.ylabel(r"Extinction factor")
[20]:
Text(0, 0.5, 'Extinction factor')
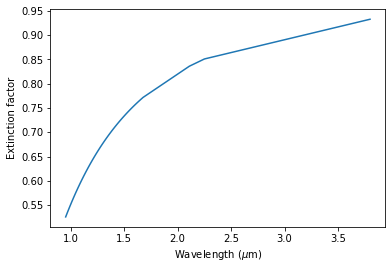
We see that as expected the effect of extinction is stronger at shorter NIR wavelengths: at 1.0 \(\mu\)m roughly only half of the flux is transmitted, while at \(L'\) band (3.8 \(\mu\)m) \(\sim\) 93% of the flux is transmitted.
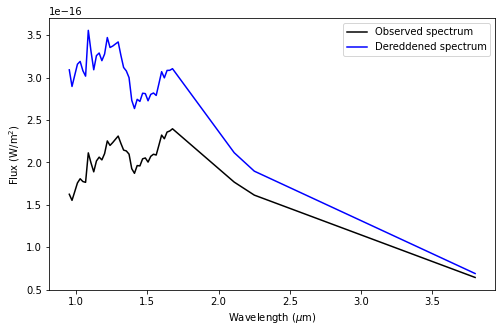
Let’s now deredden the spectrum accordingly and plot it:
[21]:
spec_dered = spec/extinc_fac
spec_dered_err = spec_err/extinc_fac
fig = plt.figure(figsize=figsize)
plt.plot(lbda, lbda*spec, 'k', label="Observed spectrum")
plt.plot(lbda, lbda*spec_dered, 'b', label="Dereddened spectrum")
plt.xlabel(r"Wavelength ($\mu$m)")
plt.ylabel(r"Flux (W/m$^2$)")
plt.legend()
[21]:
<matplotlib.legend.Legend at 0x7fd0988406d0>
3.2 Spectral indices
(Sub)stellar objects of M, L and T types have been extensively studied. Some empirical spectral indices have been proposed to estimate either their spectral type or their gravity type from the measured flux in two parts of their spectrum. Some of these spectral indices have been included in special (additional inputs are welcome). An example is provided below with the H\(_2\)O and Na indices which are relevant for a \(YJH\) spectrum.
It is important to note that different spectral indices are valid in different spectral subtype ranges! The interested reader is referred to Allers et al. (2007) and references therein.
[22]:
from special.spec_indices import spectral_idx
H2O_idx = spectral_idx(lbda, spec_dered, 'H2O-1.5', spec_dered_err, verbose=True)
Na_idx = spectral_idx(lbda, spec_dered, 'Na-1.1', spec_dered_err, verbose=True)
print("***********************")
print("H2O index: {:.3f}+-{:.3f}".format(H2O_idx[0],H2O_idx[1]))
print("Na index: {:.3f}+-{:.3f}".format(Na_idx[0],Na_idx[1]))
print("***********************")
*WARNING*: input spectrum has VERY low spectral resolution.Closest (non-overlapping) wavelengths to 1.150, 1.160, 1.134 and 1.144 mu correspond to 1.163, 1.163, 1.124 and 1.144 mu resp.Consider results with EXTRA CAUTION.
***********************
H2O index: 0.975+-0.007
Na index: 1.011+-0.004
***********************
The H\(_2\)O index can then be converted to an estimated spectral type:
[23]:
from special.spec_indices import sp_idx_to_spt, digit_to_spt
spt, spt_err = sp_idx_to_spt(H2O_idx[0], 'H2O-1.5', H2O_idx[1])
spt_str = digit_to_spt(spt, convention='Allers07')
print("SpT: {}+-{:.1f}".format(spt_str, spt_err))
SpT: M5.1+-0.8
The Na index on the other hand can be used to provide a qualitative estimate on the gravity (hence the youth) of an object:
[24]:
from special.spec_indices import sp_idx_to_gravity
sp_idx_to_gravity(Na_idx, name='Na-1.1')
The object's gravity is consistent with being very young (Na index less than 1.4; Allers et al. 2007))
4. MCMC sampler examples
Now let’s fit different models to the observed spectrum of CrA-9 b/B.
In that purpose, let’s first define some parameters that will be common to all MCMC sampler runs on this measured spectrum.
4.1. Parameters
Let’s first define the distance to the system, which will be used to scale the models (along with the companion radius):
[25]:
d_st = 153. #pc based on Gaia
The internal workings of special allow for the convolution with the spectral PSF of an IFS, or the integration over a filter transmission curve (in the case of photometric points). For these options to be considered, let’s provide the average resolving power of the IFS in YJH mode (34.5), and the name of the filenames where the filter transmission curves are encoded.
[26]:
instru_res = [34.5, 'SPHERE_IRDIS_D_K1.dat', 'SPHERE_IRDIS_D_K2.dat', 'CONICA_L_prime.dat']
Now let’s assign an index to each instrument and build a list that will tell the algorithm which instru_res values are to be assigned to which datapoints.
[27]:
n_ifs=39
instru_idx = np.array(n_ifs*[1]+[2]+[3]+[4])
In addition, we define a snippet function that can read the filter transmissions mentioned in the list. For convenience (and to allow for multiprocessing), it is defined in utils.py, and commented below FYI.
[28]:
from utils import filter_reader
# import pandas as pd
# def filter_reader(filename):
# # snippet to read filter files and return a tuple: lbda (mu), transmission
# rel_path = '../static/filters/'
# filter_path = str((par_path / rel_path).resolve())+'/'
# filt_table = pd.read_csv(filter_path+filename, sep=' ', header=0,
# skipinitialspace=True)
# keys = filt_table.keys()
# lbda_filt = np.array(filt_table[keys[0]])
# if lbda_filt.dtype == 'O':
# filt_table = pd.read_csv(filter_path+filename, sep='\t', header=0,
# skipinitialspace=True)
# keys = filt_table.keys()
# lbda_filt = np.array(filt_table[keys[0]])
# elif lbda_filt.dtype == 'int32' or lbda_filt.dtype == 'int64':
# lbda_filt = np.array(filt_table[keys[0]], dtype='float64')
# if '(AA)' in keys[0]:
# lbda_filt /=10000
# elif '(mu)' in keys[0]:
# pass
# elif '(nm)' in keys[0]:
# lbda_filt /=1000
# else:
# raise ValueError('Wavelength unit not recognised in filter file')
# trans = np.array(filt_table[keys[1]])
# return lbda_filt, trans def filter_reader(filename):
# # snippet to read filter files and return a tuple: lbda (mu), transmission
# rel_path = '../static/filters/'
# filter_path = str((par_path / rel_path).resolve())+'/'
# filt_table = pd.read_csv(filter_path+filename, sep=' ', header=0,
# skipinitialspace=True)
# keys = filt_table.keys()
# lbda_filt = np.array(filt_table[keys[0]])
# if lbda_filt.dtype == 'O':
# filt_table = pd.read_csv(filter_path+filename, sep='\t', header=0,
# skipinitialspace=True)
# keys = filt_table.keys()
# lbda_filt = np.array(filt_table[keys[0]])
# elif lbda_filt.dtype == 'int32' or lbda_filt.dtype == 'int64':
# lbda_filt = np.array(filt_table[keys[0]], dtype='float64')
# if '(AA)' in keys[0]:
# lbda_filt /=10000
# elif '(mu)' in keys[0]:
# pass
# elif '(nm)' in keys[0]:
# lbda_filt /=1000
# else:
# raise ValueError('Wavelength unit not recognised in filter file')
# trans = np.array(filt_table[keys[1]])
# return lbda_filt, trans
Let’s assemble all parameters related to the instrument(s) and filter(s) into a dictionary, including as well the spectral correlation calculated in Sec. 2:
[29]:
instru_params = {'instru_res':instru_res,
'instru_idx':instru_idx,
'instru_corr': final_sp_corr,
'filter_reader': filter_reader}
special was implemented with directly imaged substellar objects in mind. Radii and masses therefore implicitly assume Jovian units. If you prefer Solar radii for your outputs, set star to True in the following box:
[30]:
from astropy import constants as c
star = False # whether use Solar units
if star:
conv_RS = c.R_jup/c.R_sun
conv_RS
Let’s set the MCMC parameters identically for all runs shown in the next subsections (see more details in emcee documentation):
[31]:
a=2.0
nwalkers=100
niteration_min=100
niteration_limit=500 # recommended>> 1000
nproc = 2 # number of CPUs
mcmc_params = {'a':a,
'nwalkers':nwalkers,
'niteration_min':niteration_min,
'niteration_limit':niteration_limit,
'nproc':nproc}
Similarly, we adopt the autocorrelation time criterion for convergence for all MCMC runs (see more details here):
[32]:
conv_test='ac'
ac_c=50
ac_count_thr=1
conv_params = {'conv_test': conv_test,
'ac_c': ac_c,
'ac_count_thr':ac_count_thr}
Note that for the purpose of this tutorial, we set a low number of walkers and a low maximum number of iterations. It is recommended to set it large enough (e.g. >>1000, although it depends on the considered number of parameters and model) such that the convergence criterion will break the chain once it is met.
4.2. Blackbody model
This model does not involve any grid:
[33]:
grid_list=None # triggers a blackbody fit
The 2 parameters are the temperature and radius (of a sphere). It is important to label the blackbody temperature and radius ‘Tbb1’ and ‘Rbb1’, respectively. This is because special allows for the addition of an unlimited number of blackbody components. E.g. for n components, set the labels from ‘Tbb1’ to ‘Tbbn’ and from ‘Rbb1’ to ‘Rbbn’.
[34]:
labels = ('Tbb1', 'Rbb1')
units = ('K', r'$R_J$')
npar = len(labels)
Set the initial guesses and bounds accordingly. Note the requirement for a Jovian input radius.
[35]:
ini_guess = (3200., 1.3) #K and R_Jup
bounds = {'Tbb1':(2000,4000),
'Rbb1':(0.1,5)}
Again, let’s collate all model-related parameters into a dictionary:
[36]:
model_params={'grid_param_list':grid_list,
'labels':labels,
'initial_state':ini_guess,
'bounds':bounds}
The flux units of input spectra are assumed to follow the SI convention by default (W m\(^{-2} \mu\)m\(^{-1}\)). Note that special can also handle ‘cgs’ and ‘jy’. The relevant conversions will be made internally if different units are provided for observed and model spectra.
[37]:
units_obs = 'si'
Now let’s run the MCMC sampler (Warning: the next cell may take a few minutes to complete):
[38]:
from special.mcmc_sampling import mcmc_spec_sampling
res = mcmc_spec_sampling(lbda, spec, spec_err, d_st, dlbda_obs=dlbda, units_obs=units_obs,
**instru_params, **mcmc_params, **conv_params, **model_params)
Let’s convert the sampled radii into Solar radii (if requested):
[39]:
final_chain, ln_proba = res
if star:
idx_R = labels.index('Rbb1')
final_chain[:,:,idx_R] = final_chain[:,:,idx_R].copy()*conv_RS
# change units of radius
units = list(units)
units[-1] = r'$R_{\odot}$'
units = tuple(units)
Let’s inspect the walk plot:
[40]:
from special.mcmc_sampling import show_walk_plot
show_walk_plot(final_chain, labels)#, save=False,# output_dir='../datasets/',

If you wish to save the walk plot in a pdf, set save=True and provide a value for the output directory (output_dir).
Based on the walk plot, let’s define a conservative burnin factor of 0.3 and apply it to the chain:
[41]:
burnin=0.3
[42]:
cutoff = int(final_chain.shape[1]//(1/burnin))
ngood_steps = final_chain.shape[1]-cutoff
samples_flat_BB = final_chain[:, cutoff:, :].reshape((-1,npar))
#write_fits(output_dir+"isamples_flat.fits",isamples_flat)
Let’s compute the 68.27% confidence intervals on the posterior distribution, after burnin:
[43]:
from special.mcmc_sampling import confidence
bins = 100
vals, err = confidence(samples_flat_BB, labels, cfd=68.27, bins=bins,
gaussian_fit=False, weights=None,
verbose=True, save=False, bounds=bounds,
priors=None)
percentage for Tbb1: 68.70571428571428%
percentage for Rbb1: 68.51428571428573%
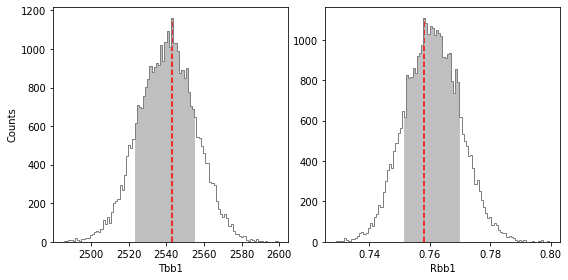
******* Results for Tbb1 *****
Confidence intervals:
Tbb1: 2542.9954443350794 [-19.820137461919785,11.892082477151689]
******* Results for Rbb1 *****
Confidence intervals:
Rbb1: 0.7582530539355073 [-0.0066963793890129075,0.011630553675653799]
If you wish to save the results in a text file, set save=True and provide a value for the output directory (output_dir).
Let’s define the most likely parameters along with their uncertainties in a numpy array:
[44]:
post_params = np.zeros([npar,3])
for i in range(npar):
post_params[i,0] = vals[labels[i]]
post_params[i,1] = err[labels[i]][0]
post_params[i,2] = err[labels[i]][1]
Now, let’s have a look at the corner plot. A couple of parameters can be fine tuned for aesthetics, e.g.:
number of significant digits for each parameter (
ndig),labels to be used in the plot for each parameter (
labels_plotcan be different to the string used inlabelsbutlabelsis used if not provided),font attributes for plot title and label,
number of bins to consider for the corner plot histograms.
[45]:
ndig = (0,2) # should have same length as labels
labels_plot = (r'$T$', r'$R$')
title_kwargs={"fontsize": 16}
label_kwargs={"fontsize": 16}
corner_bins = 20
If you wish to save the corner plot in pdf format, set save=True and provide a value to plot_name.
[46]:
from special.mcmc_sampling import show_corner_plot
#note: provide the chain before burnin, since burnin is done internally here:
show_corner_plot(final_chain, burnin=burnin, mcmc_res=post_params, units=units,
ndig=ndig, labels_plot=labels_plot, labels=labels,
#save=True, plot_name='corner_plot.pdf',
title_kwargs=title_kwargs, label_kwargs=label_kwargs, bins=corner_bins)
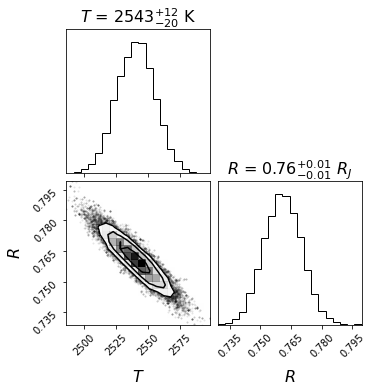
As expected, the 2 parameters are highly dependent on each other. With only 2 parameters, we also note that the estimated uncertainties on each parameter are very small. As we will see, with a more realistic model including more parameters, the uncertainties on each parameter get wider.
Let’s save the likelihood and parameter values of the most likely model for later (Sec. 5.2):
[47]:
# favoured model has highest likelihood
max_prob_BB = np.nanmax(ln_proba)
idx_max = np.unravel_index(np.nanargmax(ln_proba),shape=ln_proba.shape)
bf_params_BB = final_chain[idx_max]
bf_params_BB
[47]:
array([2.53994358e+03, 7.61007960e-01])
4.3. BT-SETTL model
Let’s now consider a grid of BT-SETTL models. In addition, let’s also consider extinction as a free parameter, since for young objects it is possible that different amounts of dust surround the companion and the central star.
In the case of BT-SETTL models, there are 2 free parameters in the grid: effective temperature and surface gravity. Let’s define the grid of parameter values covered by the models in our possession (in the models/btsettl15/ directory).
[48]:
teff_list = np.linspace(2000,4000,num=21).tolist()
logg_list = np.linspace(2.5,5.5,num=7).tolist()
grid_list = [teff_list, logg_list]
Let’s now define a snippet function that can read the input files of the grid, and provide as output a tuple of 2 vectors (1d arrays) for the wavelength and flux (in SI units, at the surface of the object). For convenience (and to allow for multiprocessing), it is defined in utils.py, and commented below FYI.
[49]:
from utils import bts_reader
# def bts_reader(params, ground=True):
# # snippet to read BT-SETTL fits files and return a tuple: lbda (mu), spec (SI)
# rel_path = '../static/btsettl15_models/'
# mod_path = str((par_path / rel_path).resolve())+'/'
# mod_units = 'cgs' # careful: input in cgs
# filename = mod_path+'btsettl15_t{:.0f}_g{:.1f}0_z-0.00_SED.txt'.format(params[0],params[1])
# lbda = []
# flux = []
# f=open(filename,"r")
# lines=f.readlines()
# for i, x in enumerate(lines):
# if i>0:
# lbda.append(float(x.split('\t')[0]))
# flux.append(float(x.split('\t')[1]))
# f.close()
# # Correct for atmospheric refraction
# flux = np.array(flux)
# lbda = np.array(lbda)
# if ground:
# # truncate at 100nm (no transmission)
# flux = flux[np.where(lbda>0.2)]
# lbda = lbda[np.where(lbda>0.2)]
# # then correct for the shift in wavelength due to atm. refraction
# nref = nrefrac(lbda*1e4)
# lbda = lbda/(1+(nref*1e-6))
# #conversion from ergs/s/cm2/um to W/m2/um
# flux = convert_F_units(flux, lbda, in_unit=mod_units, out_unit='si')
# return lbda, flux
Since the snippet function already performs the conversion to SI flux units (W m\(^{-2} \mu\)m\(^{-1}\)), we will set the model units to ‘si’. Note that ‘cgs’ units or ‘jy’ are also accepted - the conversion to match the observed flux would then be made internally.
[50]:
units_mod = 'si'
Considering the extinction A_V and photometric radius R (in Jovian radii) as free parameters, we have a total of 4 free parameters, including 2 captured by the grid: the temperature and surface gravity.
[51]:
labels = ('Teff', 'logg', 'R', 'Av')
units = ('K', '', r'$R_J$', 'mag')
npar = len(labels)
Set the initial guesses and bounds accordingly. Note the requirement for a Jovian input radius.
[52]:
ini_guess = (3100., 3.5, 1.3, 1.6)
bounds = {'Teff':(2000,4000),
'logg':(2.5,5.5),
'R':(0.1,5),
'Av':(0.,5)}
[53]:
model_params={'grid_param_list':grid_list,
'model_reader':bts_reader,
'labels':labels,
'initial_state':ini_guess,
'bounds':bounds,
'units_mod':units_mod}
In order for the MCMC sampler to run faster, all models of the grid are first resampled (at the same sampling as the measured spectrum), such that only light spectra files are opened and interpolated during the sampling. To do so, set resamp_before=True and provide a path and name where the resampled grid will be saved:
[54]:
grid_name = 'bts_resamp_grid.fits'
rel_path = '../static/bts_output/'
output_dir = str((par_path / rel_path).resolve())+'/'
resamp_before=True
Important: if a file named output_dir+grid_name already exists it will be used, instead of creating a new resampled grid (this is to save time when a given MCMC sampling is performed multiple times with different options). Make sure to change either output_dir or grid_name when considering different models or a different range of parameter values in the grid (i.e. differente grid_list).
We also set a filename where the MCMC results will be written as a pickle, such that they can be accessed without having to run again the MCMC.
[55]:
output_file = 'MCMC_results' # also written in output_dir
output_params = {'resamp_before':resamp_before,
'grid_name':grid_name,
'output_dir': output_dir,
'output_file': output_file}
Now let’s run the MCMC sampler (Warning: the next cell may take a few minutes to complete):
[56]:
from special.mcmc_sampling import mcmc_spec_sampling
res = mcmc_spec_sampling(lbda, spec, spec_err, d_st, dlbda_obs=dlbda,
**instru_params, **mcmc_params, **conv_params,
**model_params, **output_params)
Fits HDU-0 data successfully loaded. Data shape: (21, 7, 42, 2)
[57]:
final_chain, ln_proba = res
if star:
idx_R = labels.index('R')
final_chain[:,:,idx_R] = final_chain[:,:,idx_R].copy()*conv_RS
# change units of radius
units = list(units)
units[-1] = r'$R_{\odot}$'
units = tuple(units)
Let’s inspect the walk plot:
[58]:
from special.mcmc_sampling import show_walk_plot
show_walk_plot(final_chain, labels)#, save=False,# output_dir='../datasets/',
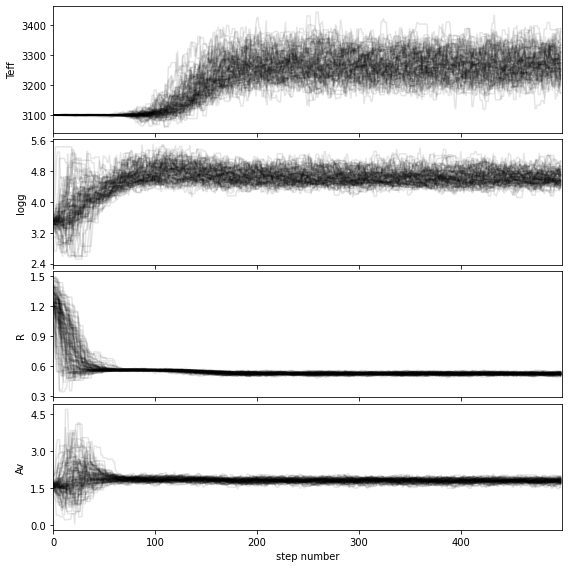
If you wish to save the walk plot in a pdf, set save=True and provide a value for the output directory (output_dir).
Based on the walk plot, let’s define a conservative burnin factor of 0.3 and apply it to the chain:
[59]:
burnin=0.3
[60]:
cutoff = int(final_chain.shape[1]//(1/burnin))
ngood_steps = final_chain.shape[1]-cutoff
samples_flat_BTS = final_chain[:, cutoff:, :].reshape((-1,npar))
#write_fits(output_dir+"isamples_flat.fits",isamples_flat)
Let’s compute the 68.27% confidence intervals on the posterior distribution, after burnin:
[61]:
from special.mcmc_sampling import confidence
bins = 100
vals, err = confidence(samples_flat_BTS, labels, cfd=68.27, bins=bins,
gaussian_fit=False, weights=None,
verbose=True, save=False, bounds=bounds,
priors=None)
percentage for Teff: 69.57714285714285%
percentage for logg: 69.28%
percentage for R: 69.23142857142857%
percentage for Av: 68.71714285714286%
******* Results for Teff *****
Confidence intervals:
Teff: 3268.537193631018 [-56.98597566100216,39.717498187971614]
******* Results for logg *****
Confidence intervals:
logg: 4.600436767117264 [-0.17341985736505894,0.21195760344618364]
******* Results for R *****
Confidence intervals:
R: 0.5296112793509183 [-0.013945448920862469,0.01056473403095648]
******* Results for Av *****
Confidence intervals:
Av: 1.8199430173064597 [-0.09084551385115014,0.09084551385115036]
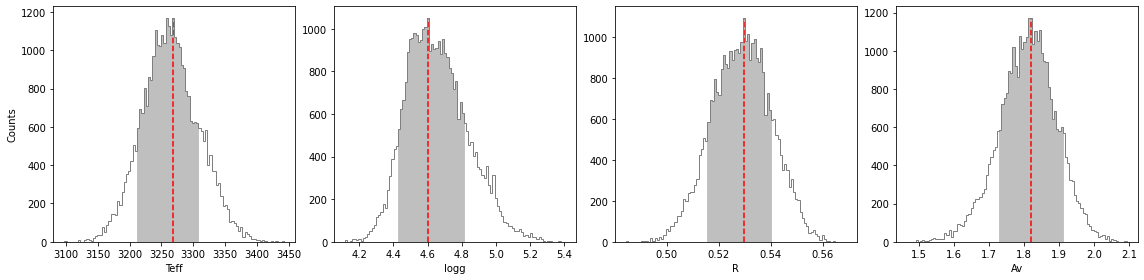
If you wish to save the results in a text file, set save=True and provide a value for the output directory (output_dir).
Let’s define the most likely parameters along with their uncertainties in a numpy array:
[62]:
post_params = np.zeros([npar,3])
for i in range(npar):
post_params[i,0] = vals[labels[i]]
post_params[i,1] = err[labels[i]][0]
post_params[i,2] = err[labels[i]][1]
Now, let’s have a look at the corner plot. A couple of parameters can be fine tuned for aesthetics, e.g.:
number of significant digits for each parameter (
ndig),labels to be used in the plot for each parameter (
labels_plotcan be different to the string used inlabelsbutlabelsis used if not provided),font attributes for plot title and label,
number of bins to consider for the corner plot histograms.
[63]:
ndig = (0,1,2,1) # should have same length as labels
labels_plot = (r'$T$', r'log($g$)', r'$R$', r'$A_V$')
title_kwargs={"fontsize": 16}
label_kwargs={"fontsize": 16}
corner_bins = 20
If you wish to save the corner plot in pdf format, set save=True and provide a value to plot_name.
[64]:
from special.mcmc_sampling import show_corner_plot
#note: provide the chain before burnin, since burnin is done internally here:
show_corner_plot(final_chain, burnin=burnin, mcmc_res=post_params,
units=units, ndig=ndig, labels_plot=labels_plot,
labels=labels, #save=True, plot_name='corner_plot.pdf',
title_kwargs=title_kwargs, label_kwargs=label_kwargs, bins=corner_bins)
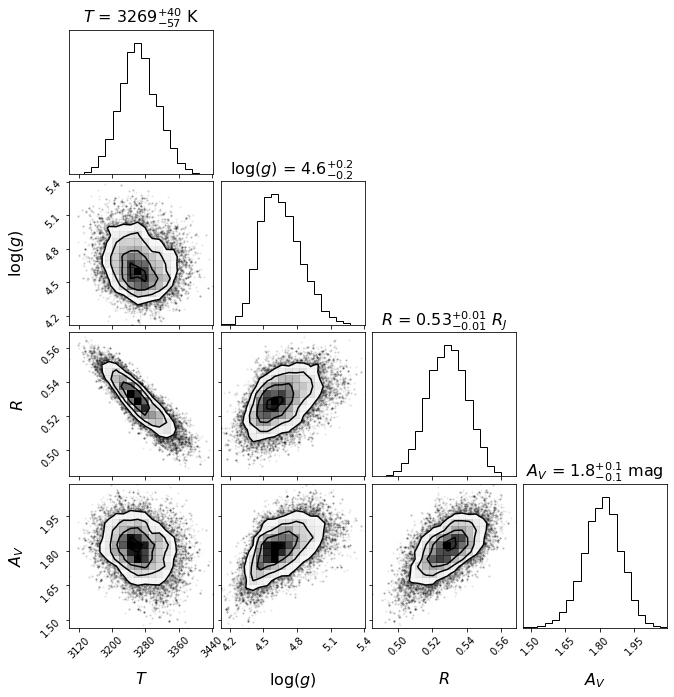
Let’s save the likelihood and parameter values of the most likely model for later (Sec. 5.2):
[65]:
# favoured model has highest likelihood
max_prob_BTS = np.nanmax(ln_proba)
idx_max = np.unravel_index(np.nanargmax(ln_proba),shape=ln_proba.shape)
bf_params_BTS = final_chain[idx_max]
bf_params_BTS
[65]:
array([3.27776316e+03, 4.51097245e+00, 5.22860539e-01, 1.77473043e+00])
The results above are puzzling: a ~3000 K effective temperature combined with a ~0.5 \(R_J\) photometric radius. See Christiaens et al. (2021) for a discussion on the different hypotheses put forward to reconcile these results.
4.4. BT-SETTL + BB model
Although this is not necessarily a good model for the NIR spectrum of CrA-9 B/b, let’s illustrate how to set up a photospheric + extra blackbody component model to further showcase the possibilities available in special.
Let’s set Gaussian priors on the temperatures of each Blackbody component, to replicate the kinds of constraints that could potentially be obtained from past works.
Note that a single extra BB component is considered here, but special does not limit the number of components you wish to incorporate in the model (just add the desired number of parameters ‘Tbb*n*’ and ‘Rbb*n*’, where n>0).
This time, we have a total of 8 free parameters, with again only 2 captured by the BT-SETTL grid:
[66]:
labels = ('Teff', 'logg', 'R', 'Av', 'Tbb1', 'Rbb1')
units = ('K', '', r'$R_J$', 'mag', 'K', r'$R_J$')
npar = len(labels)
teff_list = np.linspace(2000,4000,num=21).tolist()
logg_list = np.linspace(2.5,5.5,num=7).tolist()
grid_list = [teff_list, logg_list]
Set the initial guesses and bounds accordingly. Note the requirement for a Jovian input radius.
[67]:
ini_guess = (3100., 3.5, 1.3, 1.6, 1100, 1)
bounds = {'Teff':(2000,4000),
'logg':(2.5,5.5),
'R':(0.1,5),
'Av':(0.,5),
'Tbb1':(900,2000),
'Rbb1':(0.1,5)}
Let’s set Gaussian priors for log(\(g\)) and \(T_{bb1}\) in a dictionary. By default, uniform priors are considered for parameters not mentioned in this dictionary. Be sure to use the same dictionary labels as for parameter labels.
[68]:
priors = {'logg':(3.5,0.2),
'Tbb1':(1200,200)}
When including a blackbody component, you can ensure that the sampled solutions are physical (e.g. the BB temperatures must systematically be lower temperature than the photosphere effective temperature - assuming it comes from surrounding heated dust):
[69]:
physical=True
It is also possible to tell the model whether to apply the extinction before or after adding the blackbody component, depending on where the dust absorbing the companion signal is to be assumed with respect to the emitting dust. By default, we assume the latter (e.g. interstellar dust).
[70]:
AV_bef_bb = False
Let’s compile all model-related parameters defined above into a dictionary:
[71]:
model_params={'grid_param_list':grid_list,
'model_reader':bts_reader,
'labels':labels,
'initial_state':ini_guess,
'bounds':bounds,
'priors':priors,
'units_mod':units_mod,
'physical':physical,
'AV_bef_bb':AV_bef_bb}
In order for the MCMC sampler to run faster, all models of the grid are first resampled (at the same sampling as the measured spectrum), such that only light spectra files are opened and interpolated during the sampling. To do so, set resamp_before=True and provide a path and name where the resampled grid will be saved:
[72]:
grid_name = 'bts_bb_resamp_grid.fits'
rel_path = '../static/bts_output/'
output_dir = str((par_path / rel_path).resolve())+'/'
resamp_before=True
Important: if a file named output_dir+grid_name already exists it will be used, instead of creating a new resmpled grid (this is to save time when a given MCMC sampling is performed multiple times with different options). Make sure to change either output_dir or grid_name when considering different models or a different range of parameter values in the grid (i.e. different grid_list).
We also set a filename where the MCMC results will be written as a pickle, such that they can be accessed without having to run again the MCMC.
[73]:
output_file = 'MCMC_results_bb' # also written in output_dir
output_params = {'resamp_before':resamp_before,
'grid_name':grid_name,
'output_dir': output_dir,
'output_file': output_file}
Finally, given the larger number of parameters, let’s increase the number of iterations and walkers:
[74]:
a=2.0
nwalkers=200
niteration_min=100
niteration_limit=1000 # recommended>> 1000
nproc = 2 # number of CPUs
mcmc_params = {'a':a,
'nwalkers':nwalkers,
'niteration_min':niteration_min,
'niteration_limit':niteration_limit,
'nproc':nproc}
Now let’s run the MCMC sampler (Warning: the next cell may take a few minutes to complete):
[75]:
from special.mcmc_sampling import mcmc_spec_sampling
res = mcmc_spec_sampling(lbda, spec, spec_err, d_st, dlbda_obs=dlbda, units_obs=units_obs,
**instru_params, **mcmc_params, **conv_params, **model_params,
**output_params)
Fits HDU-0 data successfully loaded. Data shape: (21, 7, 42, 2)
[76]:
final_chain, ln_proba = res
if star:
idx_R = labels.index('R')
final_chain[:,:,idx_R] = final_chain[:,:,idx_R].copy()*conv_RS
# change units of radius
units = list(units)
units[-1] = r'$R_{\odot}$'
units = tuple(units)
Let’s inspect the walk plot:
[77]:
from special.mcmc_sampling import show_walk_plot
show_walk_plot(final_chain, labels)#, save=False,# output_dir='../datasets/',
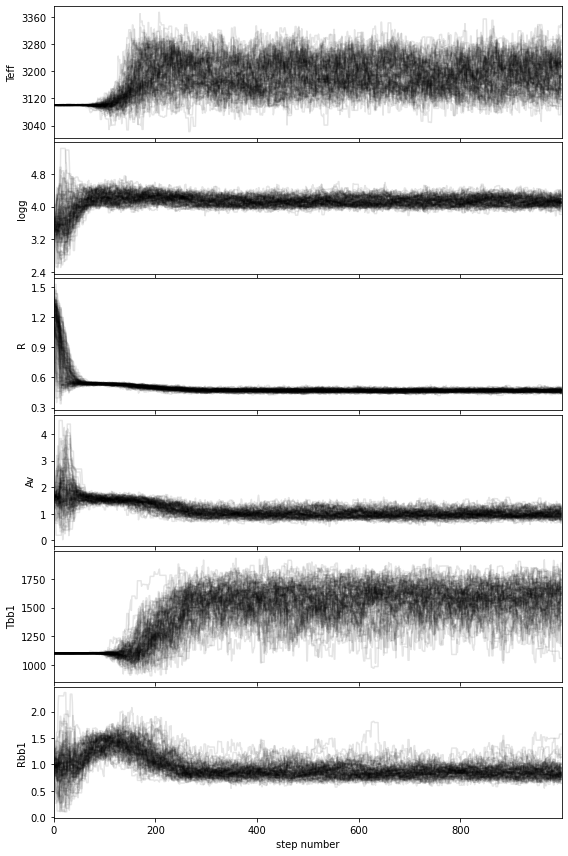
If you wish to save the walk plot in a pdf, set save=True and provide a value for the output directory (output_dir).
Based on the walk plot, let’s define a conservative burnin factor of 0.5 and apply it to the chain:
[78]:
burnin=0.5
[79]:
cutoff = int(final_chain.shape[1]//(1/burnin))
ngood_steps = final_chain.shape[1]-cutoff
samples_flat_BTS_BB = final_chain[:, cutoff:, :].reshape((-1,npar))
Let’s compute the 68.27% confidence intervals on the posterior distribution, after burnin:
[80]:
from special.mcmc_sampling import confidence
bins = 100
vals, err = confidence(samples_flat_BTS_BB, labels, cfd=68.27, bins=bins,
gaussian_fit=False, weights=None,
verbose=True, save=False, bounds=bounds,
priors=None)
percentage for Teff: 69.11699999999999%
percentage for logg: 68.27300000000001%
percentage for R: 69.448%
percentage for Av: 70.02799999999996%
percentage for Tbb1: 70.16400000000002%
percentage for Rbb1: 68.334%
******* Results for Teff *****
Confidence intervals:
Teff: 3168.8061684879867 [-32.93135392899421,67.43086756889261]
******* Results for logg *****
Confidence intervals:
logg: 4.119528892365588 [-0.12154198984640097,0.09639537125749076]
******* Results for R *****
Confidence intervals:
R: 0.47029462276193956 [-0.017623047379509982,0.01121466651423364]
******* Results for Av *****
Confidence intervals:
Av: 0.9262609450612325 [-0.1161701801150623,0.18255314018081226]
******* Results for Tbb1 *****
Confidence intervals:
Tbb1: 1625.0735030189098 [-142.87325865375692,103.45994592168563]
******* Results for Rbb1 *****
Confidence intervals:
Rbb1: 0.8454529974913282 [-0.11071662338551935,0.08466565317716201]
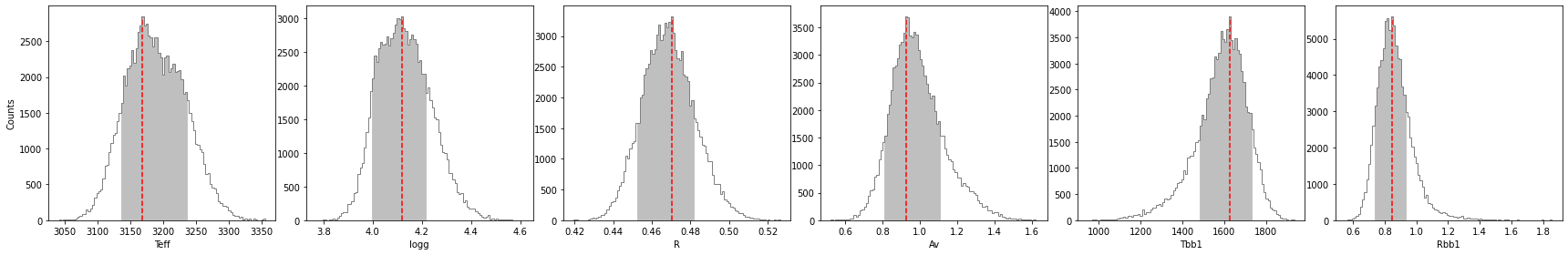
If you wish to save the results in a text file, set save=True and provide a value for the output directory (output_dir).
Let’s define the most likely parameters along with their uncertainties in a numpy array:
[81]:
post_params = np.zeros([npar,3])
for i in range(npar):
post_params[i,0] = vals[labels[i]]
post_params[i,1] = err[labels[i]][0]
post_params[i,2] = err[labels[i]][1]
Now, let’s have a look at the corner plot. A couple of parameters can be fine tuned for aesthetics, e.g.:
number of significant digits for each parameter (
ndig),labels to be used in the plot for each parameter (
labels_plotcan be different to the string used inlabelsbutlabelsis used if not provided),font attributes for plot title and label,
number of bins to consider for the corner plot histograms.
[82]:
ndig = (0,1,2,1,0,1) # should have same length as labels
labels_plot = (r'$T$', r'log($g$)', r'$R$', r'$A_V$', r'$T_{bb1}$',r'$R_{bb1}$')
title_kwargs={"fontsize": 16}
label_kwargs={"fontsize": 16}
corner_bins = 20
If you wish to save the corner plot in pdf format, set save=True and provide a value to plot_name.
[83]:
from special.mcmc_sampling import show_corner_plot
#note: provide the chain before burnin, since burnin is done internally here:
show_corner_plot(final_chain, burnin=burnin, mcmc_res=post_params, units=units,
ndig=ndig, labels_plot=labels_plot,
labels=labels, #save=True, plot_name='corner_plot.pdf',
title_kwargs=title_kwargs, label_kwargs=label_kwargs, bins=corner_bins)
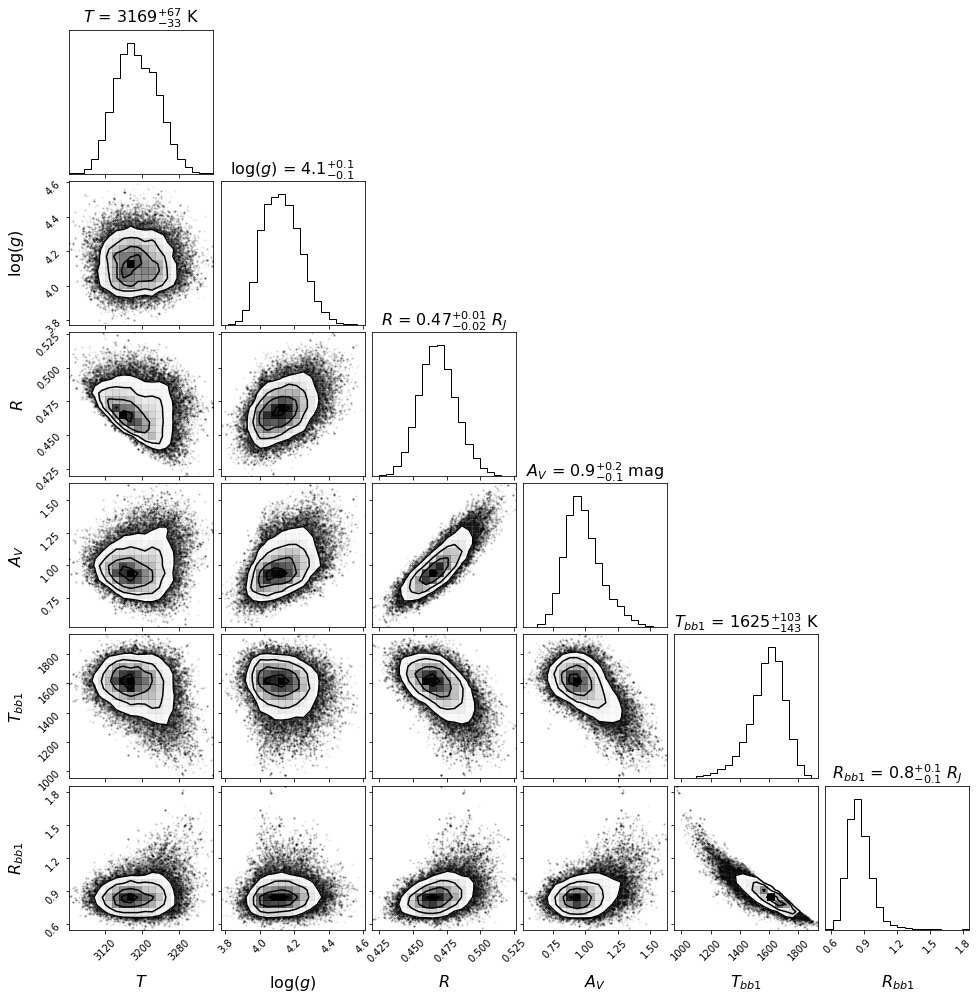
Let’s save the likelihood and parameter values of the most likely model for later (Sec. 5.2):
[84]:
# favoured model has highest likelihood
max_prob_BTS_BB = np.nanmax(ln_proba)
idx_max = np.unravel_index(np.nanargmax(ln_proba),shape=ln_proba.shape)
bf_params_BTS_BB = final_chain[idx_max]
bf_params_BTS_BB
[84]:
array([3.15611462e+03, 4.00105754e+00, 4.59765800e-01, 8.45928708e-01,
1.66216448e+03, 8.28073620e-01])
4.5. BT-SETTL model + BrG line model
In order to further showcase the possibilities available in special, let’s now consider a BT-SETTL + Brackett Gamma line model for the spectrum. Let’s also consider \(R_V\) as a free parameter, and finally consider Gaussian priors for log(\(g\)) and \(R_V\): \(\mu_{\mathrm{log}(g)}=3.5\), \(\sigma_{\mathrm{log}(g)} = 0.2\) and \(\mu_{R_V}=3.1\), \(\sigma_{R_V} = 0.2\).
This time, we have a total of 6 free parameters, including 2 captured by the BT-SETTL grid:
[85]:
labels = ('Teff', 'logg', 'R', 'Av', 'Rv', 'BrG')
units = ('K', '', r'$R_J$', 'mag', '', '')
npar = len(labels)
teff_list = np.linspace(2000,4000,num=21).tolist()
logg_list = np.linspace(2.5,5.5,num=7).tolist()
grid_list = [teff_list, logg_list]
We first provide an estimate for the possible BrG flux:
[86]:
import astropy.constants as con
F_BrG_estimate = 20000 # W m-2 at the surface (based on the estimate for the primary)
RB_est = 5 # rough estimate for the radius of the companion in R_J
input_unit = 'LogL' # select input units {'F','L','LogL'} for Flux (W/m2), Luminosity (W) or Log Solar Luminosity.
We then convert it into log Solar luminosity by setting the input_unit to ‘LogL’. Other formats are accepted, such as flux ‘F’ in SI units, or Solar luminosity ‘L’.
[87]:
if input_unit == 'F':
BrG_min = F_BrG_estimate*1e-5
BrG_max = F_BrG_estimate*1e5
elif input_unit == 'LogL':
# BrG flux is converted below into L_Sun
L_BrG_estimate = np.log10(4*np.pi*np.power(con.R_jup.value*RB_est,2)*F_BrG_estimate/con.L_sun.value)
# set min and max of BrG list of initial grid of tested values
BrG_min = L_BrG_estimate-5
BrG_max = L_BrG_estimate+5
elif input_unit == 'L':
BrG_min = F_BrG_estimate*1e-5*4*np.pi*np.power(con.R_jup.value*RB_est,2)
BrG_max = F_BrG_estimate*1e5*4*np.pi*np.power(con.R_jup.value*RB_est,2)
print(BrG_min,BrG_max)
-9.076277167361013 0.9237228326389868
Let’s set the initial guesses and bounds accordingly. Note the requirement for a Jovian input radius.
[88]:
# define list of BrG line luminosities
n_BrG_list = 23 # desired number of points in grid
BrG_list = np.linspace(L_BrG_estimate-5, L_BrG_estimate+5, n_BrG_list)
# during MCMC, sampled values will be interpolated from the grid defined above for faster calculation.
ini_guess = (3100., 3.5, 1.3, 1.6, 3.1, np.median(BrG_list))
bounds = {'Teff':(2000,4000),
'logg':(2.5,5.5),
'R':(0.1,5),
'Av':(0.,5),
'Rv':(1.,6),
'BrG': (BrG_min,BrG_max)}
Then provide details for each of the line(s), and add an entry in the em_lines and em_grid dictionaries for each spectral line you’d like to include in the model. Lines can be included in the model without necessarily fitting for their flux with MCMC (e.g. for a known emission line luminosity to be included in the model):
[89]:
em_lines = {}
em_lines['BrG'] = (2.1667, input_unit, None) # WL, unit and flux (if known). Flux should be None if to be sampled by MCMC.
em_grid = {}
em_grid['BrG'] = BrG_list.copy()
Let’s set Gaussian priors for \(R_V\) in a dictionary. By default, uniform priors are considered for parameters not mentioned in this dictionary. Be sure to use the same dictionary labels as for parameter labels.
[90]:
priors = {'logg':(3.5,0.2),
'Rv':(3.1,0.2)}
Let’s compile all model-related parameters defined above into a dictionary:
[91]:
model_params={'grid_param_list':grid_list,
'model_reader':bts_reader,
'labels':labels,
'initial_state':ini_guess,
'bounds':bounds,
'em_lines':em_lines,
'em_grid':em_grid,
'priors':priors,
'units_mod':units_mod}
In order for the MCMC sampler to run faster, all models of the grid are first resampled (at the same sampling as the measured spectrum), such that only light spectra files are opened and interpolated during the sampling. To do so, set resamp_before=True and provide a path and name where the resampled grid will be saved:
[92]:
grid_name = 'bts_BrG_resamp_grid.fits'
rel_path = '../static/bts_output/'
output_dir = str((par_path / rel_path).resolve())+'/'
resamp_before = True
Important: if a file named output_dir+grid_name already exists it will be used, instead of creating a new resmpled grid (this is to save time when a given MCMC sampling is performed multiple times with different options). Make sure to change either output_dir or grid_name when considering different models or a different range of parameter values in the grid (i.e. different grid_list).
We also set a filename where the MCMC results will be written as a pickle, such that they can be accessed without having to run again the MCMC.
[93]:
output_file = 'MCMC_results_BrG' # also written in output_dir
output_params = {'resamp_before':resamp_before,
'grid_name':grid_name,
'output_dir': output_dir,
'output_file': output_file}
Finally, given the larger number of parameters, we increase the number of iterations and walkers:
[94]:
a=2.0
nwalkers=250
niteration_min=100
niteration_limit=1500 # recommended>> 1000
nproc = 2 # number of CPUs
mcmc_params = {'a':a,
'nwalkers':nwalkers,
'niteration_min':niteration_min,
'niteration_limit':niteration_limit,
'nproc':nproc}
Now let’s run the MCMC sampler (Warning: the next cell may take a few minutes to complete):
[95]:
from special.mcmc_sampling import mcmc_spec_sampling
res = mcmc_spec_sampling(lbda, spec, spec_err, d_st, dlbda_obs=dlbda, **instru_params,
**mcmc_params, **conv_params, **model_params, **output_params)
Fits HDU-0 data successfully loaded. Data shape: (21, 7, 23, 42, 2)
[96]:
final_chain, ln_proba = res
if star:
idx_R = labels.index('R')
final_chain[:,:,idx_R] = final_chain[:,:,idx_R].copy()*conv_RS
# change units of radius
units = list(units)
units[-1] = r'$R_{\odot}$'
units = tuple(units)
Let’s inspect the walk plot:
[97]:
from special.mcmc_sampling import show_walk_plot
show_walk_plot(final_chain, labels)#, save=False,# output_dir='../datasets/',
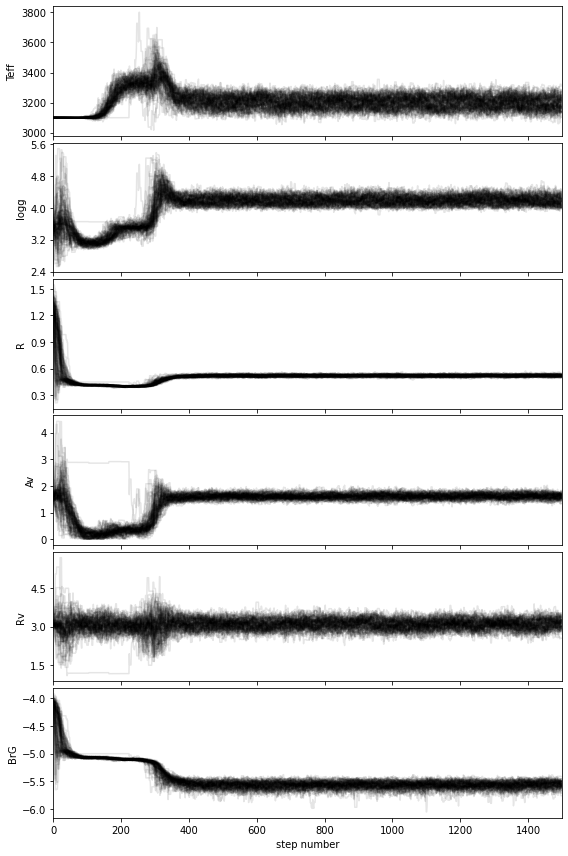
If you wish to save the walk plot in a pdf, set save=True and provide a value for the output directory (output_dir).
Based on the walk plot, let’s define a conservative burnin factor of 0.5 and apply it to the chain:
[98]:
burnin=0.5
[99]:
cutoff = int(final_chain.shape[1]//(1/burnin))
ngood_steps = final_chain.shape[1]-cutoff
samples_flat_BrG = final_chain[:, cutoff:, :].reshape((-1,npar))
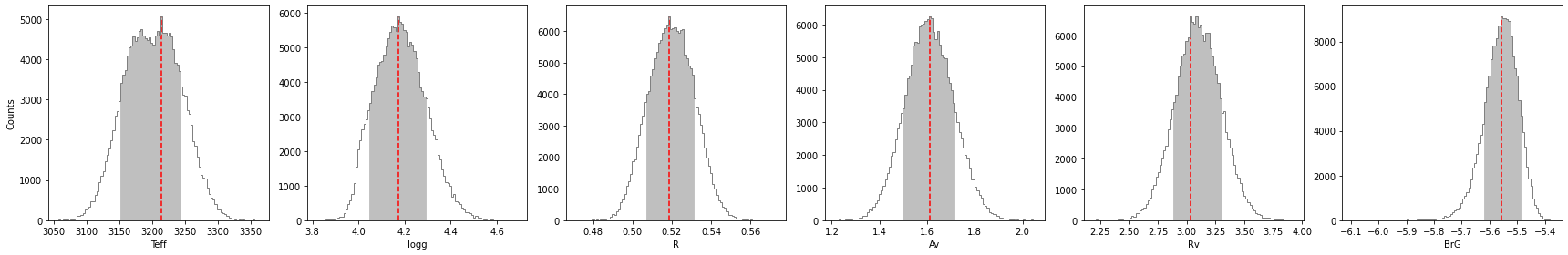
Let’s compute the 68.27% confidence intervals on the posterior distribution, after burnin:
[100]:
from special.mcmc_sampling import confidence
bins = 100
vals, err = confidence(samples_flat_BrG, labels, cfd=68.27, bins=bins, gaussian_fit=False, weights=None,
verbose=True, save=False, bounds=bounds, priors=None)
percentage for Teff: 70.03626666666666%
percentage for logg: 69.91200000000002%
percentage for R: 69.87893333333332%
percentage for Av: 69.43573333333333%
percentage for Rv: 68.2736%
percentage for BrG: 70.11466666666666%
******* Results for Teff *****
Confidence intervals:
Teff: 3214.0617331626163 [-62.31713260605284,28.8786712076826]
******* Results for logg *****
Confidence intervals:
logg: 4.172630088740946 [-0.1255693907007469,0.11690943272138554]
******* Results for R *****
Confidence intervals:
R: 0.5185647316546919 [-0.011700883183572208,0.012718351286491347]
******* Results for Av *****
Confidence intervals:
Av: 1.6110630569440505 [-0.11327637237633703,0.10488552997808998]
******* Results for Rv *****
Confidence intervals:
Rv: 3.033495340047154 [-0.14717938504144934,0.26838593742852535]
******* Results for BrG *****
Confidence intervals:
BrG: -5.557861370673404 [-0.061250222520235376,0.0684561310520273]
If you wish to save the results in a text file, set save=True and provide a value for the output directory (output_dir).
Let’s define the most likely parameters along with their uncertainties in a numpy array:
[101]:
post_params = np.zeros([npar,3])
for i in range(npar):
post_params[i,0] = vals[labels[i]]
post_params[i,1] = err[labels[i]][0]
post_params[i,2] = err[labels[i]][1]
Now, let’s have a look at the corner plot. A couple of parameters can be fine tuned for aesthetics, e.g.:
number of significant digits for each parameter (
ndig),labels to be used in the plot for each parameter (
labels_plotcan be different to the string used inlabelsbutlabelsis used if not provided),font attributes for plot title and label,
number of bins to consider for the corner plot histograms.
[102]:
ndig = (0,1,2,1,1,1) # should have same length as labels
labels_plot = (r'$T$', r'log($g$)', r'$R$', r'$A_V$', r'$R_V$', r"$\log(\frac{L_{\rm BrG}}{L_{\odot}})$")
title_kwargs={"fontsize": 16}
label_kwargs={"fontsize": 16}
corner_bins = 20
If you wish to save the corner plot in pdf format, set save=True and provide a value to plot_name.
[103]:
from special.mcmc_sampling import show_corner_plot
#note: provide the chain before burnin, since burnin is done internally here:
show_corner_plot(final_chain, burnin=burnin, mcmc_res=post_params,
units=units, ndig=ndig, labels_plot=labels_plot,
labels=labels, #save=True, plot_name='corner_plot.pdf',
title_kwargs=title_kwargs, label_kwargs=label_kwargs, bins=corner_bins)
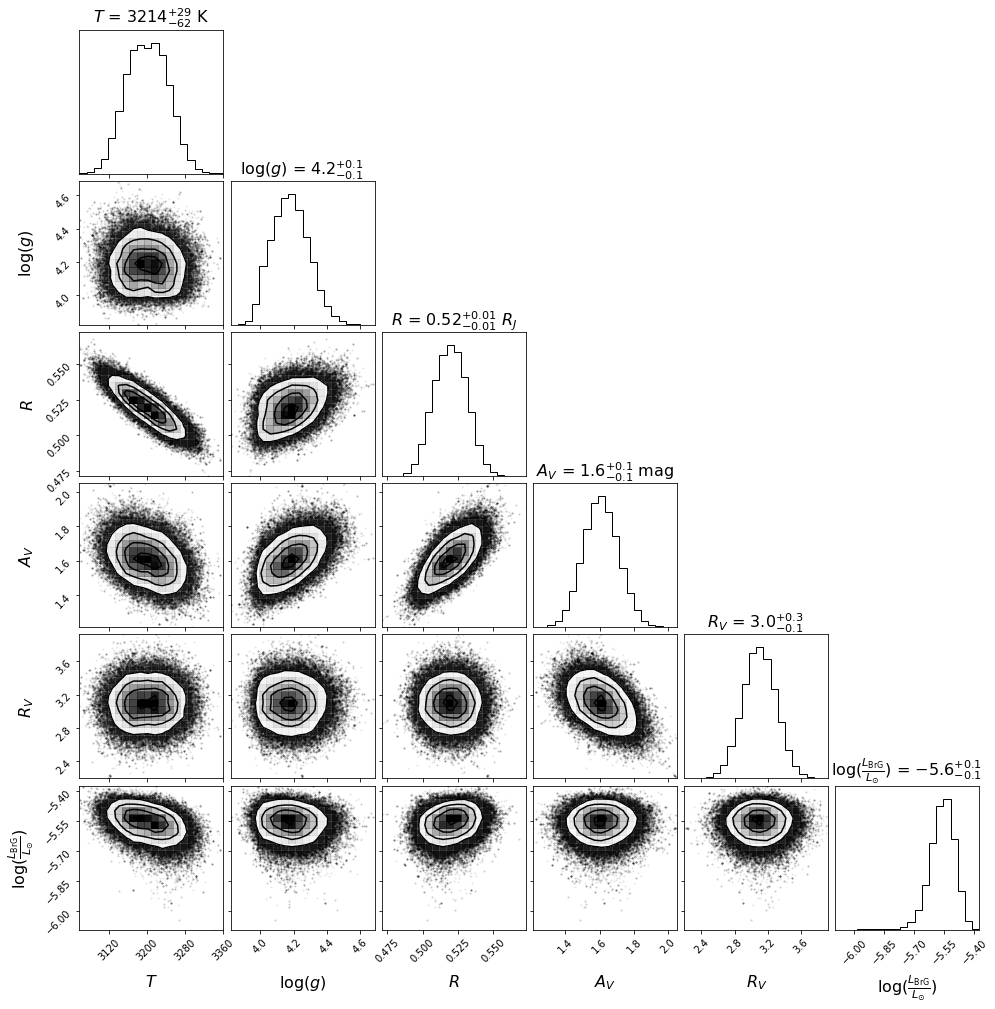
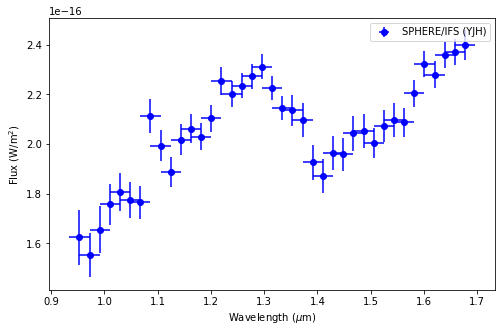
Compared to the results obtained without the inclusion of a potential BrG line (affecting the K1 photometry of the point source), we now get a lower value of log(g), more consistent with a young object. Nonetheless, the required BrG line luminosity is significant. This would likely imply the presence of more H recombination lines - which may or may not be present near ~1.09 and ~1.22 \(\mu\)m (see below).
[104]:
fig = plt.figure(figsize=figsize)
plt.errorbar(lbda[:-3], lbda[:-3]*spec[:-3], lbda[:-3]*spec_err[:-3], dlbda[:-3], 'bo',
label = 'SPHERE/IFS (YJH)')
plt.xlabel(r"Wavelength ($\mu$m)")
plt.ylabel(r"Flux (W/m$^2$)")
plt.legend()
[104]:
<matplotlib.legend.Legend at 0x7fd0502bb9a0>
Let’s save the likelihood and parameter values of the most likely model for later (Sec. 5.2):
[105]:
# favoured model has highest likelihood
max_prob_BTS_BrG = np.nanmax(ln_proba)
idx_max = np.unravel_index(np.nanargmax(ln_proba),shape=ln_proba.shape)
bf_params_BTS_BrG = final_chain[idx_max]
bf_params_BTS_BrG
[105]:
array([ 3.22149387e+03, 4.17221394e+00, 5.14884278e-01, 1.59929704e+00,
3.05956541e+00, -5.57878381e+00])
5. Comparison of results
5.1. Akaike Information Criterion
For each of the tested type of model above (Secs. 4.2 to 4.5), let’s compute the Akaike Information Criterion (AIC) in order to assess which one is likely the best model for our observations. The 4 tested models are:
Blackbody (2 parameters);
BT-SETTL + extinction (4 parameters);
BT-SETTL + 2BB + extinction (8 parameters);
BT-SETTL + BrG + extinction (6 parameters)
For each type of model, we need the maximum likelihood model among all MCMC samples, and the number of free parameters.
[106]:
nparam_list = [2, 4, 6, 6]
max_prob_list = [max_prob_BB, max_prob_BTS, max_prob_BTS_BB, max_prob_BTS_BrG]
n_models = len(nparam_list)
[107]:
from special.utils_spec import akaike
aic_list = [akaike(max_prob_list[i],nparam_list[i]) for i in range(n_models)]
aic_list
[107]:
[297.15318710404495, 166.65696588450723, 138.64392704392702, 156.765131720631]
Now let’s compute the \(\Delta\) AIC, i.e. the difference with the smallest AIC value obtained for the different models:
[108]:
min_aic = np.amin(aic_list)
daic_list = [aic_list[i]-min_aic for i in range(n_models)]
daic_list
[108]:
[158.50926006011792, 28.013038840580208, 0.0, 18.121204676703968]
The values above suggest that the most likely types of model that we considered are the BT-SETTL alone, the BT-SETTL+BB and BT-SETTL+BrG line models.
Since the difference is >>10 for the BB model, we can conclude that there is no support for that kind of model (Burnham & Anderson 2002).
5.2. Best-fit models
Let’s first assemble all the relevant parameters from the different models considered above in different lists:
[109]:
lab_models = ['BB', 'BT-SETTL', 'BT-SETTL + BB', 'BT-SETTL + BrG line']
bf_params = [bf_params_BB, bf_params_BTS, bf_params_BTS_BB, bf_params_BTS_BrG]
all_samples_flat = [samples_flat_BB, samples_flat_BTS, samples_flat_BTS_BB, samples_flat_BrG]
all_labels = [('Tbb1', 'Rbb1'),
('Teff', 'logg', 'R', 'Av'),
('Teff', 'logg', 'R', 'Av', 'Tbb1', 'Rbb1'),
('Teff', 'logg', 'R', 'Av', 'Rv', 'BrG')]
all_grid_lists = [None, grid_list, grid_list, grid_list]
all_em_lines = [{}, {}, {}, em_lines]
all_em_grids = [{}, {}, {}, em_grid]
all_ndig = [(0,2),
(0,1,2,1),
(0,1,2,1,0,1),
(0,1,2,1,1,1)]
all_units = [('K', r'$R_J$'),
('K', '', r'$R_J$', 'mag'),
('K', '', r'$R_J$', 'mag', 'K', r'$R_J$'),
('K', '', r'$R_J$', 'mag', '', '')]
n_models = len(lab_models)
[110]:
bf_params
[110]:
[array([2.53994358e+03, 7.61007960e-01]),
array([3.27776316e+03, 4.51097245e+00, 5.22860539e-01, 1.77473043e+00]),
array([3.15611462e+03, 4.00105754e+00, 4.59765800e-01, 8.45928708e-01,
1.66216448e+03, 8.28073620e-01]),
array([ 3.22149387e+03, 4.17221394e+00, 5.14884278e-01, 1.59929704e+00,
3.05956541e+00, -5.57878381e+00])]
Now for each type of model considered above, let’s now generate the spectrum corresponding to the most likely parameters inferred with the MCMC. By providing lbda_obs to make_model_from_params, the returned model will be resampled to match the spectral resolution of the measured spectrum. If provided, the instrumental resolving power and/or photometric filter(s) will also be used. If lbda_obs is left to None, the model is returned at the native resolution of the grid used as input.
Let’s leverage this to have both high-res and resampled spectra.
[111]:
from special.model_resampling import make_model_from_params
bf_models = []
bf_models_hr = []
for nn in range(n_models):
bf_params_nn = tuple(bf_params[nn])
lbda_model, flux_model = make_model_from_params(bf_params_nn,
all_labels[nn],
all_grid_lists[nn],
d_st,
model_reader=bts_reader,
em_lines=all_em_lines[nn],
em_grid=all_em_grids[nn],
units_obs=units_obs,
units_mod=units_mod,
interp_order=1,
lbda_obs=lbda,
dlbda_obs=dlbda,
instru_res=instru_res,
instru_idx=instru_idx,
filter_reader=filter_reader)
bf_models.append([lbda_model, flux_model])
if nn > 0:
lbda_model_hr, flux_model_hr = make_model_from_params(bf_params_nn,
all_labels[nn],
all_grid_lists[nn],
d_st,
model_reader=bts_reader,
em_lines=all_em_lines[nn],
em_grid=all_em_grids[nn],
units_obs=units_obs,
units_mod=units_mod,
interp_order=1)
bf_models_hr.append([lbda_model_hr, flux_model_hr])
For models not based on a grid, lbda_obs is a mandatory parameter, hence the avoidance of the BB model for high-res models. Let’s use the output lbda_model_hr from BT-SETTL models, to add a high-res spectrum for the BB model as well.
[112]:
lbda_model_hr, flux_model_hr = make_model_from_params(tuple(bf_params[0]),
all_labels[0],
all_grid_lists[0],
d_st,
model_reader=bts_reader,
em_lines=all_em_lines[0],
em_grid=all_em_grids[0],
units_obs=units_obs,
units_mod=units_mod,
interp_order=1,
lbda_obs=bf_models_hr[-1][0])
bf_models_hr = [[lbda_model_hr, flux_model_hr]]+bf_models_hr
Now crop the model spectra to the same wavelength range as the measured spectrum:
[113]:
from special.utils_spec import find_nearest
for nn in range(n_models):
lbda_model_hr, flux_model_hr = bf_models_hr[nn]
# LOAD BEST FIT MODEL
idx_ini = find_nearest(lbda_model_hr,0.99*lbda[0], constraint='floor')
idx_fin = find_nearest(lbda_model_hr,1.01*lbda[-1], constraint='ceil')
lbda_model_hr = lbda_model_hr[idx_ini:idx_fin+1]
flux_model_hr = flux_model_hr[idx_ini:idx_fin+1]
dlbda_hr = np.mean(lbda_model_hr[1:]-lbda_model_hr[:-1])
bf_models_hr[nn] = [lbda_model_hr, flux_model_hr]
Now let’s plot the best-fit models at their native resolution (first panel) and after resampling (second panel):
[114]:
fig, axes = plt.subplots(2,1,figsize=(11,12))
# plot options
cols = ['k', 'r', 'b', 'c', 'y', 'm'] # colors of different models
maj_tick_sp = 0.5 # WL spacing for major ticks
min_tick_sp = maj_tick_sp / 5. # WL spacing for minor ticks
# Titles
axes[0].set_title("Best-fit models (original resolution)")
axes[1].set_title("Best-fit models (resampled)")
# Plot measured spectrum
axes[0].errorbar(lbda, lbda*spec,
lbda*spec_err, fmt=cols[0]+'o',
label='Measured spectrum')
axes[1].errorbar(lbda, lbda*spec,
lbda*spec_err, fmt=cols[0]+'o',
label='Measured spectrum')
# Set axes labels
axes[0].set_xlabel(r"Wavelength ($\mu$m)")
axes[0].set_ylabel(r"$\lambda F_{\lambda}$ (W m$^{-2} \mu$m$^{-1}$)")
axes[1].set_xlabel(r"Wavelength ($\mu$m)")
axes[1].set_ylabel(r"$\lambda F_{\lambda}$ (W m$^{-2} \mu$m$^{-1}$)")
# Plot best-fit models
for i in range(2):
if i == 0:
bf_models_i = bf_models_hr
else:
bf_models_i = bf_models
for nn in range(n_models):
lbda_model, flux_model = bf_models_i[nn]
lab_str = '{0} = {1:.{2}f} {3}'
if nn==0 or nn ==1:
show_labs = range(len(bf_params[nn]))
elif nn==2:
show_labs = range(len(bf_params[nn])-1)
else:
show_labs = list(range(len(bf_params[nn])-2))
lab_str_list = [lab_str.format(all_labels[nn][j], bf_params[nn][j], all_ndig[nn][j], all_units[nn][j]) for j in show_labs]
sep = ', '
label = "{} model: {}".format(lab_models[nn], sep.join(lab_str_list))
axes[i].plot(lbda_model, lbda_model*flux_model, cols[1+nn], linewidth=2,
alpha=0.5, label=label)
min_tick = lbda[0]-dlbda[0]/2-((lbda[0]-dlbda[0]/2)%0.2)
max_tick = lbda[-1]+dlbda[-1]/2+(0.2-((lbda[-1]+dlbda[-1]/2)%0.2))
major_ticks1 = np.arange(min_tick,max_tick,maj_tick_sp)
minor_ticks1 = np.arange(min_tick,max_tick,min_tick_sp)
axes[i].set_xticks(major_ticks1)
axes[i].set_xticks(minor_ticks1, minor = True)
axes[i].tick_params(which = 'both', direction = 'in')
axes[i].legend(loc='best')
plt.show()
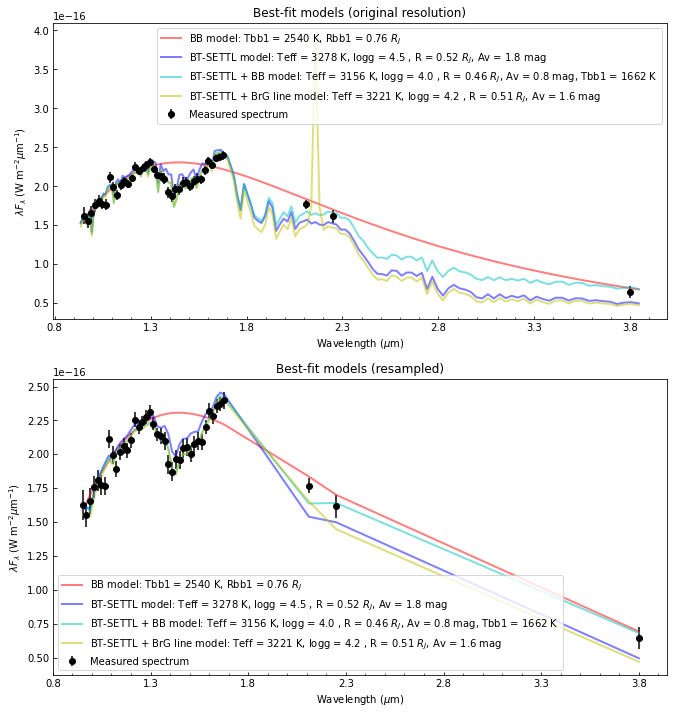
Visually, it looks like a combination of a \(T_{\rm eff}\sim 3200\) K BT-SETTL model, an extra black-body component (\(T_{\rm eff}\sim 1700\) K) and several emission lines may be able to reproduce the entire observed spectrum. However, one has to be cautious of not overfitting the data …
5.3. Models from the posterior distribution
It can be useful to plot a large number of models randomly picked from the posterior distribution in order to have an idea of the uncertainty in different parts of the spectrum. Let’s consider 200 samples from the posterior distribution:
[115]:
n_samp = 200
[116]:
import random
all_plot_samples = []
for nn in range(n_models):
print("################## Model {} ##################".format(lab_models[nn]))
samp_flux = []
lbda_samp = None # use native model resolution (set to `lbda` for measured spectrum sampling)
if nn == 0:
# set the wavelength sampling for
lbda_samp=bf_models_hr[-1][0]
for ii in range(n_samp):
# draw a random integer
idx = random.randint(0, len(all_samples_flat[nn]))
param_samp = tuple(all_samples_flat[nn][idx])
samp_lbda, tmp = make_model_from_params(param_samp, all_labels[nn], all_grid_lists[nn], d_st,
model_reader=bts_reader, em_lines=all_em_lines[nn],
em_grid=all_em_grids[nn], units_obs=units_obs,
units_mod=units_mod, interp_order=1, lbda_obs=lbda_samp)
samp_flux.append(tmp)
# Crop to same range as measurements
idx_ini = find_nearest(samp_lbda,0.99*lbda[0], constraint='floor')
idx_fin = find_nearest(samp_lbda,1.01*lbda[-1], constraint='ceil')
samp_lbda = samp_lbda[idx_ini:idx_fin+1]
samp_fluxes = [samp_flux[ii][idx_ini:idx_fin+1] for ii in range(n_samp)]
all_plot_samples.append([samp_lbda,samp_fluxes])
################## Model BB ##################
################## Model BT-SETTL ##################
################## Model BT-SETTL + BB ##################
################## Model BT-SETTL + BrG line ##################
/Users/valentin/GitHub/special/special/model_resampling.py:284: RuntimeWarning: overflow encountered in power
flux_ratio_ext = np.power(10., -extinc_curve/2.5)
Now let’s plot the samples:
[117]:
fig, axes = plt.subplots(4,1,figsize=(11,24))
# plot options
cols = ['k', 'r', 'b', 'c', 'y', 'm'] # colors of different models
maj_tick_sp = 0.5 # WL spacing for major ticks
min_tick_sp = maj_tick_sp / 5. # WL spacing for minor ticks
# Titles
for nn in range(n_models):
axes[nn].set_title("Sample models from posterior distribution ({})".format(lab_models[nn]))
# Set axes labels
axes[nn].set_xlabel(r"Wavelength ($\mu$m)")
axes[nn].set_ylabel(r"$\lambda F_{\lambda}$ (W m$^{-2} \mu$m$^{-1}$)")
# Options depending on model
if nn==0 or nn ==1:
show_labs = range(len(bf_params[nn]))
elif nn==2:
show_labs = range(len(bf_params[nn])-1)
else:
show_labs = list(range(len(bf_params[nn])-2))
# Plot sample models
lbda_samp, fluxes_model = all_plot_samples[nn]
for i in range(n_samp):
flux_model = fluxes_model[i]
if i == 0:
lab_str = '{0} = {1:.{2}f} {3}'
lab_str_list = [lab_str.format(all_labels[nn][j], bf_params[nn][j], all_ndig[nn][j], all_units[nn][j]) for j in show_labs]
sep = ', '
label = "{} model samples".format(lab_models[nn])#: {}".format(lab_models[nn], sep.join(lab_str_list))
else:
label = None
axes[nn].plot(lbda_samp, lbda_samp*flux_model, cols[1+nn], linewidth=2, alpha=0.1, label=label)
min_tick = lbda[0]-dlbda[0]/2-((lbda[0]-dlbda[0]/2)%0.2)
max_tick = lbda[-1]+dlbda[-1]/2+(0.2-((lbda[-1]+dlbda[-1]/2)%0.2))
major_ticks1 = np.arange(min_tick,max_tick,maj_tick_sp)
minor_ticks1 = np.arange(min_tick,max_tick,min_tick_sp)
axes[nn].set_xticks(major_ticks1)
axes[nn].set_xticks(minor_ticks1, minor = True)
axes[nn].tick_params(which = 'both', direction = 'in')
# Plot measured spectrum
axes[nn].errorbar(lbda, lbda*spec,
lbda*spec_err, fmt=cols[0]+'o',
label='Measured spectrum')
axes[nn].legend(loc='best')
#plt.savefig("Sample_models.pdf", bbox_inches='tight')
plt.show()
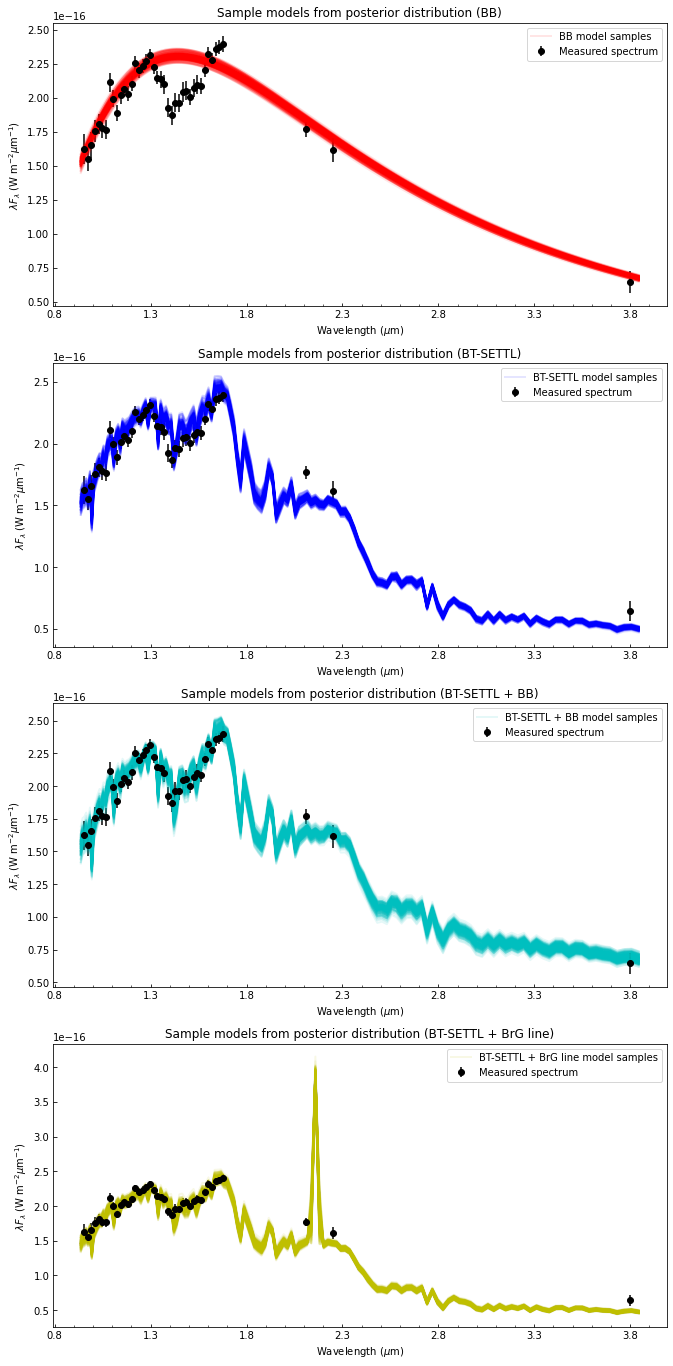
6. Nested sampler examples
The procedure to set the nested sampler routines is similar to what we saw with the MCMC, as most parameters of mcmc_spec_sampling and nested_spec_sampling are identical.
The main differences are the sampler, method, npoints or dlogz parameters for the nested samplers:
the
samplercan be set to'ultranest'or'nestle'.in the case of
'nestle', themethodcan be set to'classic'(MCMC; Skilling 2004),'single'(single ellipsoid; Mutherjee et al. 2006) or'multi'(original multinest implementation as in Feroz et al. 2009, with conservative ellipsoid splitting).npointscorresponds to the number of active live points (minimum number in the case of UltraNest);dlogzis the target evidence uncertainty. The stopping criterion is log(z + z_est) - log(z) < dlogz. There are additional criteria for UltraNest.
Other sampler-specific parameters (e.g. decline_factor and rstate for nestle, or frac_remain and dKL for UltraNest) can be provided as kwargs. The interested reader is referred to the documentation of UltraNest and nestle for details on how to appropriately set the values of the different parameters.
Below we show three examples of nested sampler runs, using nestle and ultranest, with the BT-SETTL grid, considering A_V as a free parameter. Let’s first set the parameters as in Sec. 4.3:
[118]:
from utils import bts_reader
teff_list = np.linspace(2000,4000,num=21).tolist()
logg_list = np.linspace(2.5,5.5,num=7).tolist()
grid_list = [teff_list, logg_list]
ini_guess = (3100., 3.5, 1.3, 1.6)
labels = ('Teff', 'logg', 'R', 'Av')
labels_plot = (r'$T$', r'log($g$)', r'$R$', r'$A_V$')
units = ('K', '', r'$R_J$', 'mag')
ndig = (0,1,2,1) # number of significant digits
units_mod = 'si'
bounds = {'Teff':(2000,4000),
'logg':(2.5,5.5),
'R':(0.1,5),
'Av':(0.,5)}
model_params={'grid_param_list':grid_list,
'model_reader':bts_reader,
'labels':labels,
'bounds':bounds,
'units_mod':units_mod}
grid_name='bts_resamp_grid.fits'
rel_path = '../static/bts_output/'
output_dir = str((par_path / rel_path).resolve())+'/'
resamp_before=True
output_params = {'resamp_before':resamp_before,
'grid_name':grid_name,
'output_dir': output_dir}
6.1. Nestle - single ellipsoid method
Now let’s set the parameters that are specific to the nested sampler:
[119]:
sampler='nestle'
method='single'
npoints=1500
dlogz=0.1
[120]:
nested_params = {'sampler':sampler,
'method':method,
'npoints':npoints,
'dlogz':dlogz}
Now let’s run the nested sampler. Warning: the next cell may take up to a few minutes to complete - reduce the number of active points for a faster calculation.
[121]:
from special.nested_sampling import nested_spec_sampling
res = nested_spec_sampling(ini_guess, lbda, spec, spec_err, d_st, dlbda_obs=dlbda, **instru_params,
**nested_params, **model_params, **output_params)
Fits HDU-0 data successfully loaded. Data shape: (21, 7, 42, 2)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2023-05-08 16:59:27
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Prior bounds on parameters:
Teff [2000,4000]
logg [2.5,5.5]
R [0.1,5]
Av [0.0,5]
Using 1500 active points
Total running time:
Running time: 0:06:05.177341
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Now let’s plot the results:
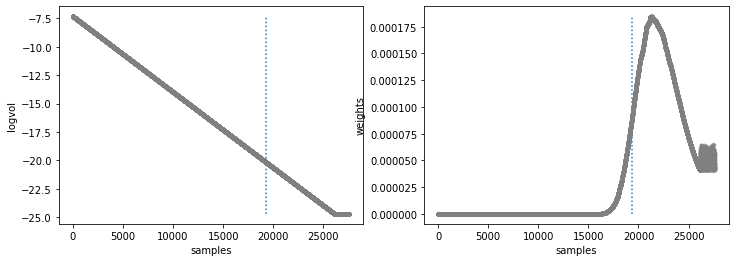
[122]:
from special.nested_sampling import show_nestle_results
final_res = show_nestle_results(res, labels, method=method, burnin=0.7, bins=None, cfd=68.27,
units=units, ndig=ndig, labels_plot=labels_plot, save=False,
output_dir='./', plot=True)
niter: 26105
ncall: 95512
nsamples: 27605
logz: -94.391 +/- 0.093
h: 13.064
Natural log of prior volume and Weight corresponding to each sample
Walk plots before the burnin
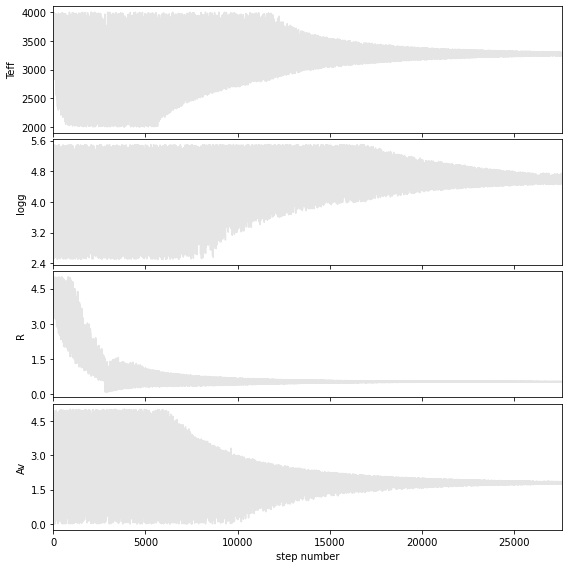
Walk plots after the burnin
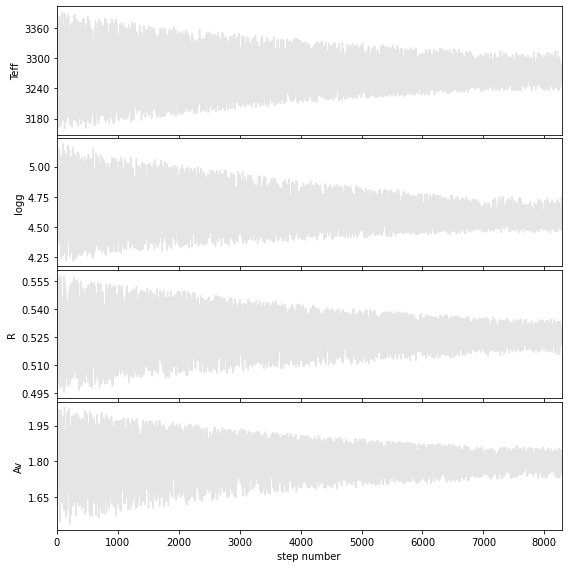
Weighted mean +- sqrt(covariance)
Teff = 3270 +/- 41
logg = 4.6 +/- 0.2
R = 0.53 +/- 0.01
Av = 1.8 +/- 0.1
Hist bins = 91
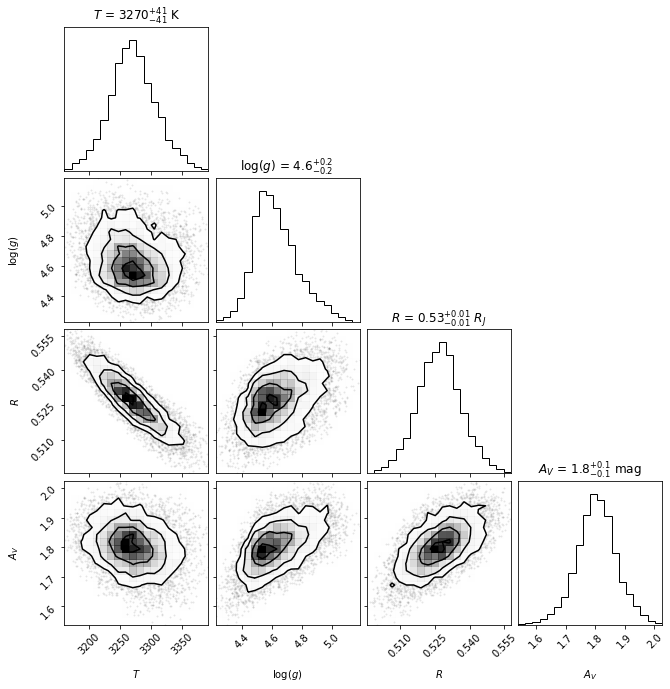
Confidence intervals
percentage for Teff: 68.69325372637766%
percentage for logg: 69.00875209816401%
percentage for R: 68.58988627363547%
percentage for Av: 69.12226646174167%
******* Results for Teff *****
Confidence intervals:
Teff: 3265.6231426509084 [-47.32906217538448,49.887389860540225]
Gaussian fit results:
Teff: 3270.232216747061 +-37.552482976221874
******* Results for logg *****
Confidence intervals:
logg: 4.566973411065405 [-0.1219748077333822,0.24925286797691104]
Gaussian fit results:
logg: 4.629821414822097 +-0.1497690355250289
******* Results for R *****
Confidence intervals:
R: 0.52729907409839 [-0.01196794055372663,0.010600175919015076]
Gaussian fit results:
R: 0.5268292282938275 +-0.009526056906226689
******* Results for Av *****
Confidence intervals:
Av: 1.7820752344394402 [-0.05625272255452329,0.10446934188697177]
Gaussian fit results:
Av: 1.8038934043192472 +-0.06874061336251147
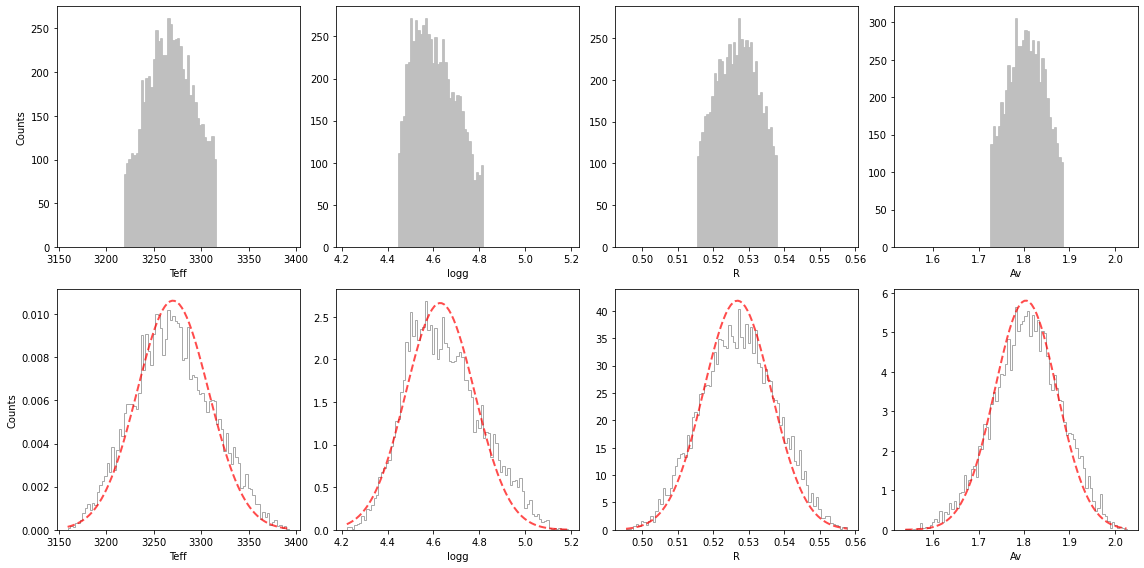
The best-fit parameters, with uncertainty are:
[123]:
for p in range(len(labels)):
fmt = "{{:.{0}f}}".format(ndig[p]).format
line = r"{0}: {1}+/-{2} {3}"
print(line.format(labels[p],fmt(final_res[p][0]),fmt(final_res[p][1]), units[p]))
Teff: 3270+/-41 K
logg: 4.6+/-0.2
R: 0.53+/-0.01 $R_J$
Av: 1.8+/-0.1 mag
6.2. Nestle - multi-ellipsoid method
Let’s adapt the method and number of active points:
[124]:
sampler='nestle'
method='multi'
npoints=2000
dlogz=0.5
[125]:
nested_params = {'sampler':sampler,
'method':method,
'npoints':npoints,
'dlogz':dlogz}
Now let’s run the nested sampler. Warning: the next cell may take up to a few minutes to complete - reduce the number of active points for a faster calculation.
[126]:
from special.nested_sampling import nested_spec_sampling
res = nested_spec_sampling(ini_guess, lbda, spec, spec_err, d_st, dlbda_obs=dlbda, **instru_params,
**nested_params, **model_params, **output_params)
Fits HDU-0 data successfully loaded. Data shape: (21, 7, 42, 2)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2023-05-08 17:05:34
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Prior bounds on parameters:
Teff [2000,4000]
logg [2.5,5.5]
R [0.1,5]
Av [0.0,5]
Using 2000 active points
Total running time:
Running time: 0:07:13.804959
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Now let’s plot the results:
[127]:
from special.nested_sampling import show_nestle_results
final_res = show_nestle_results(res, labels, method=method, burnin=0.7, bins=None, cfd=68.27,
units=units, ndig=ndig, labels_plot=labels_plot, save=False,
output_dir='./', plot=True)
niter: 31211
ncall: 110849
nsamples: 33211
logz: -94.207 +/- 0.080
h: 12.896
Natural log of prior volume and Weight corresponding to each sample
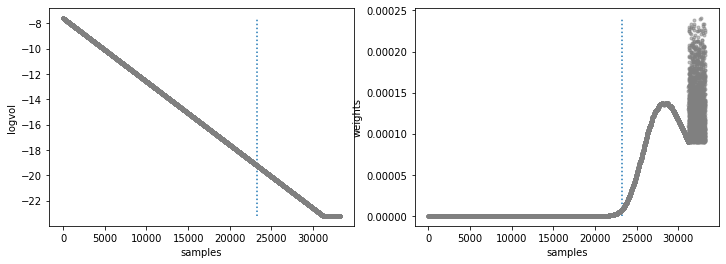
Walk plots before the burnin
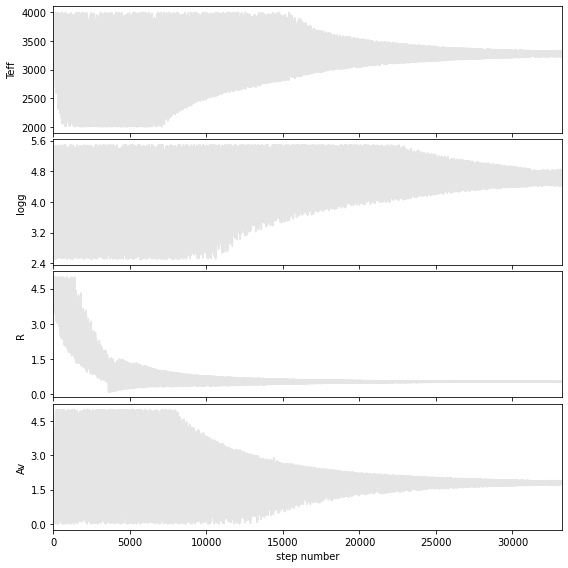
Walk plots after the burnin
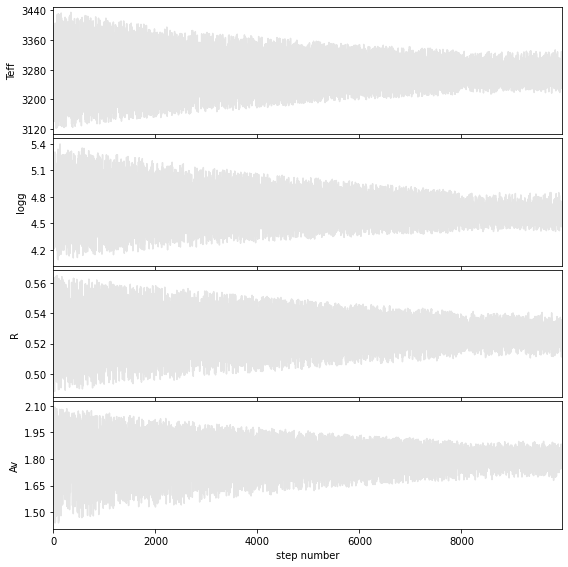
Weighted mean +- sqrt(covariance)
Teff = 3270 +/- 44
logg = 4.7 +/- 0.2
R = 0.53 +/- 0.01
Av = 1.8 +/- 0.1
Hist bins = 99
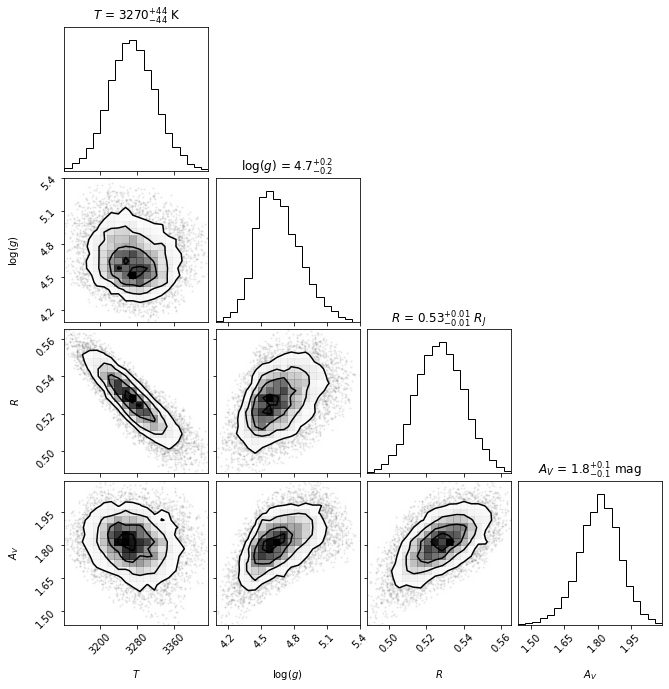
Confidence intervals
percentage for Teff: 69.72033635934162%
percentage for logg: 68.38961119720196%
percentage for R: 68.88926242249346%
percentage for Av: 69.4935601917666%
******* Results for Teff *****
Confidence intervals:
Teff: 3269.203121254572 [-45.727599259344515,45.72759925934406]
Gaussian fit results:
Teff: 3270.466306068884 +-50.82823562458199
******* Results for logg *****
Confidence intervals:
logg: 4.518232629957659 [-0.0862444516965839,0.27200173227384017]
Gaussian fit results:
logg: 4.666203996451627 +-0.20315435230105794
******* Results for R *****
Confidence intervals:
R: 0.5259084225783466 [-0.012026642989035552,0.012802555439941132]
Gaussian fit results:
R: 0.5272967777306088 +-0.012667103230549065
******* Results for Av *****
Confidence intervals:
Av: 1.825295179162057 [-0.10888022904255945,0.06928741848162878]
Gaussian fit results:
Av: 1.8077748841054169 +-0.0940790048831327
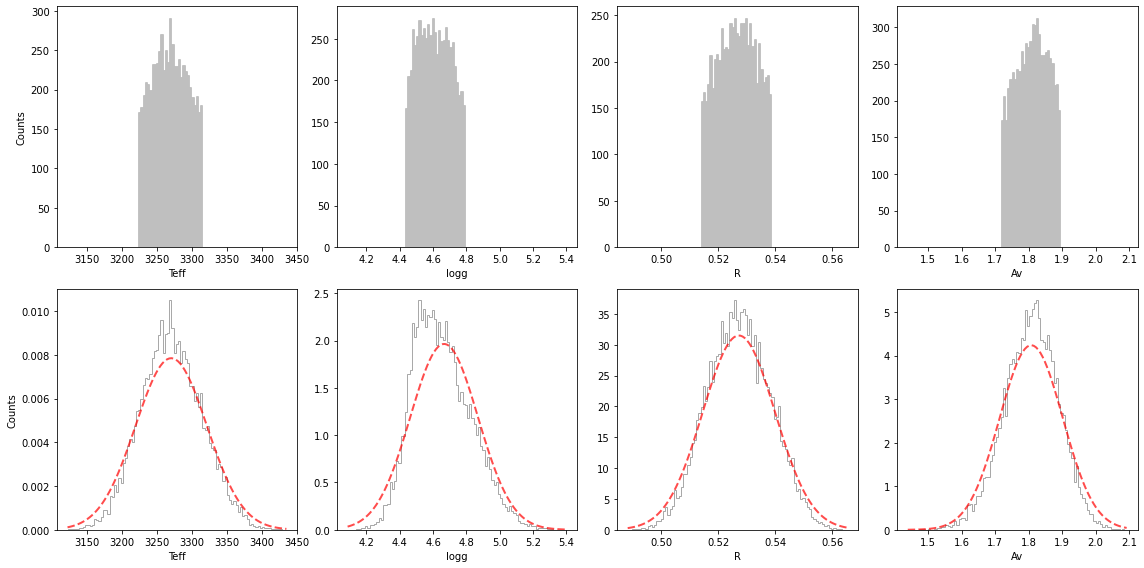
We see that for the case of the spectrum of CrA-9B/b with a BT-SETTL + extinction model, even when we allow for multiple nests, the highest likelihood area in the parameter space still corresponds to an individual ellipsoid.
6.3. UltraNest
Let’s adapt the sampler and number of active points:
[128]:
sampler='ultranest'
npoints=1000
dlogz=0.2
[129]:
nested_params = {'sampler':sampler,
'method':method,
'npoints':npoints,
'dlogz':dlogz}
Now let’s run the nested sampler. Warning: the next cell may take up to a few minutes to complete - reduce the number of active points for a faster calculation.
[130]:
from special.nested_sampling import nested_spec_sampling
res = nested_spec_sampling(ini_guess, lbda, spec, spec_err, d_st, dlbda_obs=dlbda, **instru_params,
**nested_params, **model_params, **output_params)
Fits HDU-0 data successfully loaded. Data shape: (21, 7, 42, 2)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2023-05-08 17:12:50
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Prior bounds on parameters:
Teff [2000,4000]
logg [2.5,5.5]
R [0.1,5]
Av [0.0,5]
Using 1000 active points
[ultranest] Resuming from 20543 stored points
[ultranest] Explored until L=-8e+01 [-79.4751..-79.4751]*| it/evals=19500/49465 eff=inf% N=1000 00 0 00 0
[ultranest] Likelihood function evaluations: 49465
[ultranest] Writing samples and results to disk ...
[ultranest] Writing samples and results to disk ... done
[ultranest] logZ = -94.36 +- 0.08149
[ultranest] Effective samples strategy satisfied (ESS = 5363.6, need >400)
[ultranest] Posterior uncertainty strategy is satisfied (KL: 0.46+-0.05 nat, need <0.50 nat)
[ultranest] Evidency uncertainty strategy is satisfied (dlogz=0.08, need <0.2)
[ultranest] logZ error budget: single: 0.11 bs:0.08 tail:0.01 total:0.08 required:<0.20
[ultranest] done iterating.
Total running time:
Running time: 0:00:23.576898
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Now let’s plot the results:
[131]:
from special.nested_sampling import show_ultranest_results
del model_params["bounds"] # unused for ultranest results plot
del instru_params["instru_corr"] # unused for ultranest results plot
final_res = show_ultranest_results(res, bins=None, cfd=68.27, save=False, plot=True,
units=units, ndig=ndig, labels_plot=labels_plot,
lbda_obs=lbda, spec_obs=spec, spec_obs_err=spec_err, dist=d_st,
**model_params, **instru_params)
logZ = -94.355 +- 0.125
single instance: logZ = -94.355 +- 0.114
bootstrapped : logZ = -94.361 +- 0.125
tail : logZ = +- 0.010
insert order U test : converged: True correlation: inf iterations
Teff : 3084 │ ▁ ▁▁▁▁▁▁▂▂▃▄▅▆▇▇▇▇▇▅▅▄▃▃▂▁▁▁▁▁▁▁▁▁▁ ▁ │3494 3270 +- 44
logg : 4.00 │ ▁▁▁▁▁▁▁▁▁▂▃▄▆▆▇▇▇▆▆▆▅▅▄▃▃▃▂▂▁▁▁▁▁▁▁▁▁ │5.38 4.66 +- 0.18
R : 0.480 │ ▁▁▁▁▁▁▁▁▁▁▂▂▃▅▆▆▇▇▇▇▆▆▅▅▃▃▃▂▁▁▁▁▁▁▁ ▁ │0.574 0.527 +- 0.011
Av : 1.368 │ ▁ ▁▁▁▁▁▁▁▁▁▁▂▃▃▅▆▇▇▇▇▇▆▅▃▃▂▁▁▁▁▁▁ ▁ │2.172 1.808 +- 0.083
Confidence intervals
Hist bins = 98
percentage for Teff: 69.05748319588042%
percentage for logg: 68.61098073412055%
percentage for R: 68.49686732810508%
percentage for Av: 69.70769181174423%
******* Results for Teff *****
Confidence intervals:
Teff: 3258.5838073052732 [-39.84428271142815,60.81495782270622]
******* Results for logg *****
Confidence intervals:
logg: 4.577720445280006 [-0.1451559643056921,0.2368334154461298]
******* Results for R *****
Confidence intervals:
R: 0.5256796962813602 [-0.011878513577833094,0.013779075750286363]
******* Results for Av *****
Confidence intervals:
Av: 1.829998138709031 [-0.11273860280808967,0.06263255711560567]
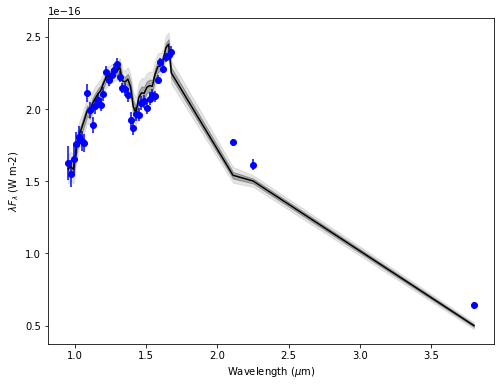
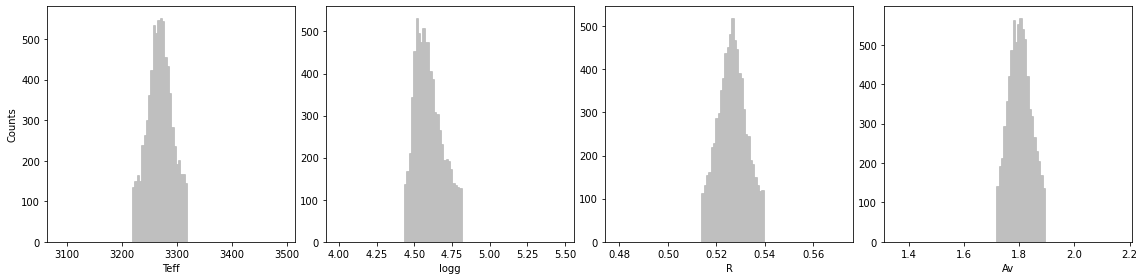
percentage for Teff: 69.05748319588042%
percentage for logg: 68.61098073412055%
percentage for R: 68.49686732810508%
percentage for Av: 69.70769181174423%
******* Results for Teff *****
Confidence intervals:
Teff: 3258.5838073052732 [-39.84428271142815,60.81495782270622]
Gaussian fit results:
Teff: 3269.932232046592 +-47.65653344442819
******* Results for logg *****
Confidence intervals:
logg: 4.577720445280006 [-0.1451559643056921,0.2368334154461298]
Gaussian fit results:
logg: 4.644950396124002 +-0.19952209766044599
******* Results for R *****
Confidence intervals:
R: 0.5256796962813602 [-0.011878513577833094,0.013779075750286363]
Gaussian fit results:
R: 0.5270857975433556 +-0.011702451604743036
******* Results for Av *****
Confidence intervals:
Av: 1.829998138709031 [-0.11273860280808967,0.06263255711560567]
Gaussian fit results:
Av: 1.8050380170168783 +-0.08960446777712383
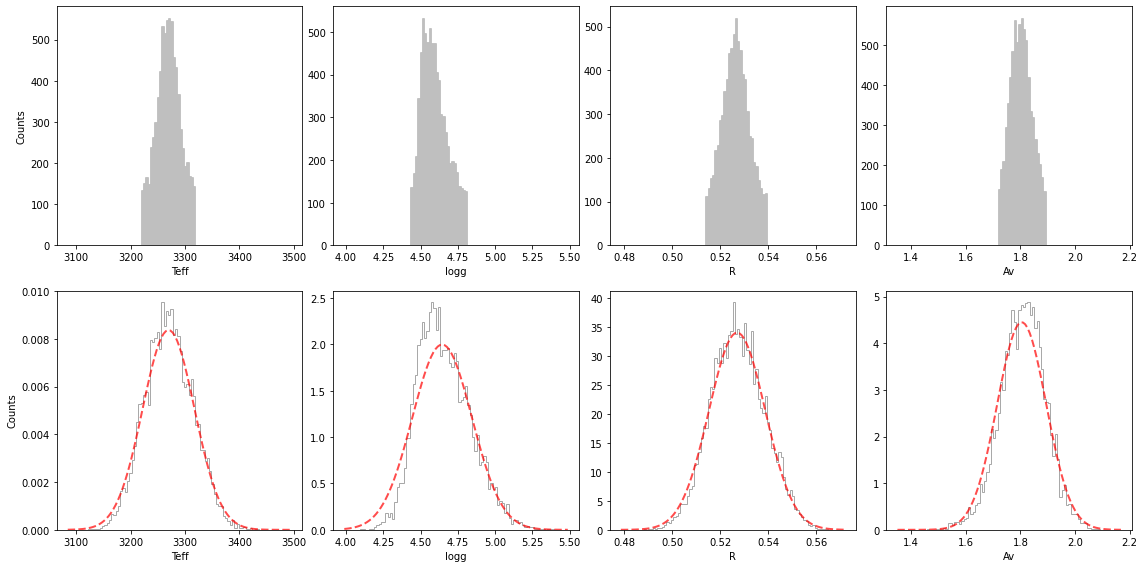
Teff = 3270 +/- 7
logg = 4.6 +/- 0.4
R = 0.53 +/- 0.11
Av = 1.8 +/- 0.3
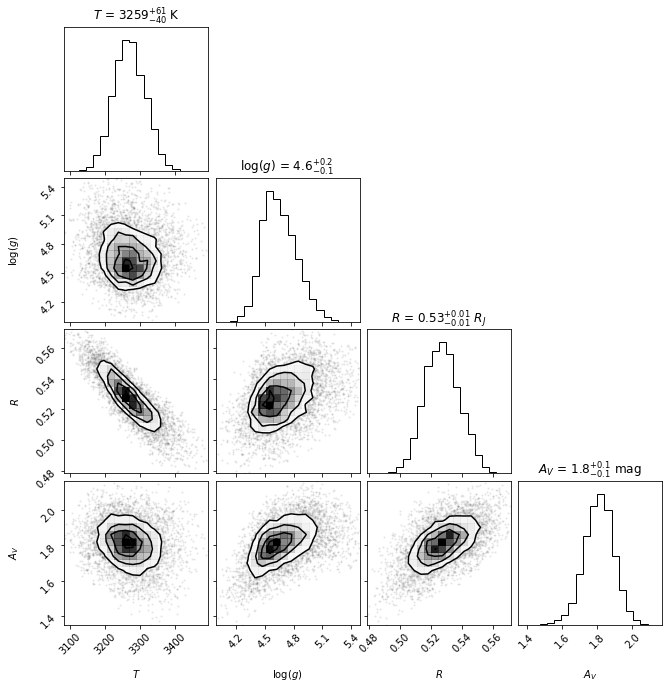
We see that for the case of the spectrum of CrA-9B/b with a BT-SETTL + extinction model, UltraNest also finds that the highest likelihood area in the parameter space corresponds to an individual ellipsoid.
7. Best-fit template spectrum
In this section, we will find the best-fit template spectra to our measured spectrum in the Montreal Spectral Library of observed M-L-T type low-mass objects. Since the template spectra from this library typically do not cover wavelengths longward of the K band, we will discard the NACO L’ point from the spectrum, just for the sake of this exercise:
[132]:
lbda_crop = lbda[:-1]
spec_crop = spec[:-1]
spec_err_crop = spec_err[:-1]
dlbda_crop = dlbda[:-1]
Let’s define the parameters associated to the instrument(s) which acquired the spectrum (again we discard the NACO measurement):
[133]:
instru_res_crop = instru_res[:-1]
instru_idx_crop = instru_idx[:-1]
final_sp_corr_crop = final_sp_corr[:-1,:-1]
instru_params = {'instru_res':instru_res_crop,
'instru_idx':instru_idx_crop,
'instru_corr':final_sp_corr_crop,
'filter_reader':filter_reader}
For this exercice, we have downloaded the MSL library following the instructions on the page of J. Gagné: https://jgagneastro.com/the-montreal-spectral-library/
Below, we then set the folder in which the template spectra placed, and their extension (‘.fits’, for automatic identification of spectra in that folder). We also define the snippet function tmp_reader to read the template spectra in the utils.py file, loaded below.
[134]:
from utils import tmp_reader
rel_path = '../static/MSL/'
inpath_models = str((par_path / rel_path).resolve())+'/'
tmp_endswith='.fits'
tmp_params = {'tmp_reader':tmp_reader,
'lib_dir':inpath_models,
'tmp_endswith':tmp_endswith}
FYI it is commented below:
[135]:
# def tmp_reader(tmp_name, verbose=False):
# rel_path = '../static/MSL/'
# inpath_tmp = str((par_path / rel_path).resolve())+'/'
# if verbose:
# print("opening {}".format(tmp_name))
# spec_model = open_fits(inpath_tmp+tmp_name, verbose=False,
# output_verify='ignore')
# lbda_model = spec_model[0]
# flux_model = spec_model[1]
# flux_model_err = spec_model[2]
# return lbda_model, flux_model, flux_model_err
Let’s assume we are interested in the top 3 most similar template spectra:
[136]:
n_best=3
Let’s use the Nelder-Mead algorithm to find the optimal flux scaling factor and \(A_V\) to be applied to each template. Additional options for the simplex can be set with simplex_options (example above) - these will be provided to the scipy.minimize function.
[137]:
search_mode = 'simplex' #'grid'
simplex_options = {'xatol': 1e-6, 'fatol': 1e-6, 'maxiter': 1000, 'maxfev': 5000}
AV_range = (-3, 7, 0.1) # min, max, step. Note: in simplex mode, min and max values are only used to set a chi=np.inf outside of these bounds.
search_opt = {'search_mode':search_mode,
'simplex_options':simplex_options,
#'scale_range':(0.2, 5, 0.05),# min, max, step
'ext_range':AV_range}
Alternatively, one can set search_mode to 'grid', which will simply use a grid search to find the optimal \(A_V\) and flux scaling factors to be applied to the template spectra for comparison to the measured spectrum. This is slower but avoids the risk of a failed minimization. In this case, both the AV_range and scale_range parameters have to be set.
Some template spectra may be corrupted or unusable for this specific spectrum (e.g. wrong wavelength coverage), let’s force the algorithm to continue instead of raising an error.
[138]:
force_continue=True
Some template spectra will have a sufficient wavelength coverage (Y to K), but with missing points (NaN values) in between bands (e.g. between J and H bands, or H and K bands). For a given template spectrum to be considered valid, it is recommeded to set a minimum threshold in terms of number of non-NaN resampled points. The risk of not doing that is to end up with best-fit template spectra with very few - but very well fitting - points minimizing our \(\chi_r^2\) metric.
Let’s set this threshold to cover >80% of our observed spectrum, i.e. 34 points:
[139]:
min_npts = 34
Now let’s find the best-fit templates:
[140]:
from special.template_fit import best_fit_tmp
final_res = best_fit_tmp(lbda_crop, spec_crop, spec_err_crop, dlbda_obs=dlbda_crop, n_best=n_best,
**tmp_params, **search_opt, **instru_params, force_continue=force_continue,
verbosity=1, nproc=1, min_npts=min_npts)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2023-05-08 17:22:55
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
424 template spectra will be tested.
****************************************
Unsufficient number of good points (33 < 34). Tmp discarded.
1/424 (2MASSJ0428+1839_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0313-2447_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
2/424 (2MASSJ0313-2447_NIR_Flamingos-2_Gagne2015c.fits) FAILED
3/424: SIMPJ2215+2110_NIR_SpeX_Robert2016, chi_r^2 = 43.1, ndof=39
Unsufficient number of good points (33 < 34). Tmp discarded.
4/424 (2MASSJ2353-1844A_NIR_SpeX_Gagne2015c.fits) FAILED
Wavelength range of template SIMPJ1150+0520_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
5/424 (SIMPJ1150+0520_NIR_NIRI_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ2238+2304_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
6/424 (SIMPJ2238+2304_NIR_SpeX_Robert2016.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2353A_merge' / [astropy.io.fits.card]
7/424: 2MASSJ1045-2819_NIR_SpeX_Gagne2015c, chi_r^2 = 5.4, ndof=33
8/424: SIMPJ1501-1831_NIR_SpeX_Robert2016, chi_r^2 = 10.5, ndof=39
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ2251-6811_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
9/424 (2MASSJ2251-6811_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2251-6811_ALLCOMB_merge' / [astropy.io.fits.card]
10/424: SIMPJ0429+0607_NIR_GNIRS_Robert2016, chi_r^2 = 34.6, ndof=39
11/424: SIMPJ1058+1339_NIR_SIMON_Robert2016, chi_r^2 = 22.4, ndof=39
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ0752-7947_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
12/424 (2MASSJ0752-7947_NIR_Flamingos-2_Gagne2015c.fits) FAILED
13/424: 2MASSJ0443+0002_NIR_SpeX_Gagne2015c, chi_r^2 = 14.7, ndof=39
Wavelength range of template SIMPJ2148+2239_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
14/424 (SIMPJ2148+2239_NIR_NIRI_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2202-5605_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
15/424 (2MASSJ2202-5605_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2202-5605_ALLCOMB_merge' / [astropy.io.fits.card]
16/424: SIMPJ0951-1343_NIR_SIMON_Robert2016, chi_r^2 = 33.6, ndof=39
17/424: 2MASSJ1119-3917_NIR_SpeX_Gagne2015c, chi_r^2 = 2.6, ndof=33
18/424: SIMPJ1627+0836_NIR_SpeX_Robert2016, chi_r^2 = 11.3, ndof=39
19/424: 2MASSJ0344+0716B_NIR_SpeX_Gagne2015c, chi_r^2 = 3.3, ndof=33
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ1510-1147_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.43)mu?
20/424 (SIMPJ1510-1147_NIR_SpeX_Robert2016.fits) FAILED
21/424: SIMPJ1415+2635_NIR_SpeX_Robert2016, chi_r^2 = 14.5, ndof=39
Wavelength range of template SIMPJ1613-0747_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
22/424 (SIMPJ1613-0747_NIR_NIRI_Robert2016.fits) FAILED
Unsufficient number of good points (30 < 34). Tmp discarded.
23/424 (2MASSJ2240+0532_NIR_SpeX_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2240+0532_merge' / [astropy.io.fits.card]
24/424: SIMPJ2322-1407_NIR_SpeX_Robert2016, chi_r^2 = 26.6, ndof=39
Wavelength range of template SIMPJ1225-1013_NIR_GNIRS_Robert2016.fits (1.00, 2.52)mu too short compared to that of observed spectrum (0.95, 2.25)mu
25/424 (SIMPJ1225-1013_NIR_GNIRS_Robert2016.fits) FAILED
No indices match the constraint (ceil w.r.t 2.36)
Issue with resampling of template SIMPJ2318-1301_NIR_SIMON_Robert2016.fits. Does the wavelength range extend far enough (0.80, 2.27)mu?
26/424 (SIMPJ2318-1301_NIR_SIMON_Robert2016.fits) FAILED
27/424: SIMPJ1217+0708_NIR_GNIRS_Robert2016, chi_r^2 = 14.4, ndof=35
28/424: SIMPJ0858+2710_NIR_GNIRS_Robert2016, chi_r^2 = 115.6, ndof=35
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0120-5200_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
29/424 (2MASSJ0120-5200_NIR_Flamingos-2_Gagne2015c.fits) FAILED
30/424: SIMPJ0150+3827_NIR_SpeX_Robert2016, chi_r^2 = 30.7, ndof=39
Unsufficient number of good points (31 < 34). Tmp discarded.
31/424 (2MASSJ0103-2805B_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0041-5621_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
32/424 (2MASSJ0041-5621_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Wavelength range of template SIMPJ1756+2815_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
33/424 (SIMPJ1756+2815_NIR_NIRI_Robert2016.fits) FAILED
34/424: SIMPJ1031+3349_NIR_SpeX_Robert2016, chi_r^2 = 15.3, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2041-3506_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
35/424 (2MASSJ2041-3506_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2219-6828_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
36/424 (2MASSJ2219-6828_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2041-3506_ALLCOMB_merge' / [astropy.io.fits.card]
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J22191486-6828018_ALLCOMB_merge' / [astropy.io.fits.card]
37/424: SIMPJ1211+0406_NIR_SIMON_Robert2016, chi_r^2 = 18.2, ndof=39
38/424: SIMPJ0148+3712_NIR_SpeX_Robert2016, chi_r^2 = 17.2, ndof=39
39/424: SIMPJ1308+0818_NIR_SpeX_Robert2016, chi_r^2 = 9.1, ndof=39
40/424: SIMPJ0851+1043_NIR_GNIRS_Robert2016, chi_r^2 = 201.1, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0226-5327_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
41/424 (2MASSJ0226-5327_NIR_Flamingos-2_Gagne2015c.fits) FAILED
42/424: 2MASSJ0335+2342_NIR_SpeX_Gagne2015c, chi_r^2 = 7.8, ndof=39
43/424: SIMPJ2150-0412_NIR_SIMON_Robert2016, chi_r^2 = 24.8, ndof=39
Wavelength range of template SIMPJ1755+3618_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
44/424 (SIMPJ1755+3618_NIR_NIRI_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0541-0217_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
45/424 (2MASSJ0541-0217_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ0042+1142_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
46/424 (2MASSJ0042+1142_NIR_SpeX_Gagne2015c.fits) FAILED
47/424: SIMPJ0417-1838_NIR_SpeX_Robert2016, chi_r^2 = 12.3, ndof=39
48/424: SIMPJ0947+0617_NIR_SIMON_Robert2016, chi_r^2 = 8.2, ndof=39
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2244-3045_merge' / [astropy.io.fits.card]
49/424: 2MASSJ2244-3045_NIR_SpeX_Gagne2015c, chi_r^2 = 13.0, ndof=35
Wavelength range of template SIMPJ1414+0107_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
50/424 (SIMPJ1414+0107_NIR_NIRI_Robert2016.fits) FAILED
51/424: 2MASSJ0758+15301A_NIR_SpeX_Gagne2015c, chi_r^2 = 3.1, ndof=32
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0038-6403_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
52/424 (2MASSJ0038-6403_NIR_Flamingos-2_Gagne2015c.fits) FAILED
53/424: SIMPJ1058+1339_NIR_GNIRS_Robert2016, chi_r^2 = 36.0, ndof=39
54/424: SIMPJ0429+0607_NIR_SIMON_Robert2016, chi_r^2 = 24.5, ndof=39
55/424: 2MASSJ1411-2119_NIR_SpeX_Gagne2015c, chi_r^2 = 9.5, ndof=39
56/424: 2MASSJ0856-1342_NIR_SpeX_Gagne2015c, chi_r^2 = 10.1, ndof=39
57/424: 2MASSJ1207-3900_NIR_SpeX_Gagne2015c, chi_r^2 = 19.4, ndof=39
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ0418+1121_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.43)mu?
58/424 (SIMPJ0418+1121_NIR_SpeX_Robert2016.fits) FAILED
59/424: 2MASSJ0337-3709_NIR_SpeX_Gagne2015c, chi_r^2 = 3.1, ndof=33
60/424: SIMPJ2155+2345_NIR_SpeX_Robert2016, chi_r^2 = 15.3, ndof=39
61/424: SIMPJ2249-1628_NIR_SpeX_Robert2016, chi_r^2 = 25.4, ndof=39
62/424: SIMPJ1411+2948_NIR_SpeX_Robert2016, chi_r^2 = 39.4, ndof=39
63/424: SIMPJ1453-2744_NIR_GNIRS_Robert2016, chi_r^2 = 94.9, ndof=39
64/424: 2MASSJ0046+0715_NIR_SpeX_Gagne2015c, chi_r^2 = 13.5, ndof=32
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ2335-3401_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
65/424 (2MASSJ2335-3401_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0309-3014_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
66/424 (2MASSJ0309-3014_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0019-6226_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
67/424 (2MASSJ0019-6226_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2335-3401_ALLCOMB_merge' / [astropy.io.fits.card]
68/424: 2MASSJ0404+2616B_NIR_SpeX_Gagne2015c, chi_r^2 = 8.1, ndof=32
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ1547-1626B_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
69/424 (2MASSJ1547-1626B_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1227-0636_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
70/424 (2MASSJ1227-0636_NIR_Flamingos-2_Gagne2015c.fits) FAILED
71/424: 2MASSJ1948+5944B_NIR_SpeX_Gagne2015c, chi_r^2 = 3.5, ndof=35
Unsufficient number of good points (33 < 34). Tmp discarded.
72/424 (2MASSJ2011-5048_NIR_FIRE_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0259-4232_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
73/424 (2MASSJ0259-4232_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'W2011-5048_tc.fits' / [astropy.io.fits.card]
74/424: 2MASSJ1153-3015_NIR_SpeX_Gagne2015c, chi_r^2 = 2.7, ndof=33
75/424: SIMPJ2303+3150_NIR_SpeX_Robert2016, chi_r^2 = 70.2, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1156-4043_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
76/424 (2MASSJ1156-4043_NIR_Flamingos-2_Gagne2015c.fits) FAILED
77/424: SIMPJ1211+0406_NIR_GNIRS_Robert2016, chi_r^2 = 107.9, ndof=36
78/424: 2MASSJ1035-2058_NIR_SpeX_Gagne2015c, chi_r^2 = 3.3, ndof=33
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ0314+1603_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
79/424 (2MASSJ0314+1603_NIR_SpeX_Gagne2015c.fits) FAILED
80/424: SIMPJ1317+0448_NIR_SpeX_Robert2016, chi_r^2 = 14.4, ndof=39
81/424: 2MASSJ0001+1535_NIR_FIRE_Gagne2015c, chi_r^2 = 20.9, ndof=39
82/424: SIMPJ1343-1216_NIR_SIMON_Robert2016, chi_r^2 = 27.4, ndof=39
2MASSJ0039+1330B_NIR_SpeX_Gagne2015c.fits could not be opened. Corrupt file?
83/424 (2MASSJ0039+1330B_NIR_SpeX_Gagne2015c.fits) FAILED
84/424: 2MASSJ1013-1706A_NIR_SpeX_Gagne2015c, chi_r^2 = 3.0, ndof=39
85/424: SIMPJ0417+1634_NIR_SIMON_Robert2016, chi_r^2 = 14.4, ndof=39
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ0422+1033_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.43)mu?
86/424 (SIMPJ0422+1033_NIR_SpeX_Robert2016.fits) FAILED
Unsufficient number of good points (31 < 34). Tmp discarded.
87/424 (2MASSJ1846+5246B_NIR_SpeX_Gagne2015c.fits) FAILED
88/424: 2MASSJ0602-1413_NIR_SpeX_Gagne2015c, chi_r^2 = 12.2, ndof=39
89/424: 2MASSJ1106-3715_NIR_SpeX_Gagne2015c, chi_r^2 = 10.4, ndof=33
90/424: 2MASSJ0527+0007B_NIR_SpeX_Gagne2015c, chi_r^2 = 3.1, ndof=33
91/424: 2MASSJ1148-2836_NIR_Flamingos-2_Gagne2015c, chi_r^2 = 15.1, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2244-6650_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
92/424 (2MASSJ2244-6650_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2244-6650_ALLCOMB_merge' / [astropy.io.fits.card]
93/424: SIMPJ1039-1904_NIR_SpeX_Robert2016, chi_r^2 = 13.0, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1133-7807_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
94/424 (2MASSJ1133-7807_NIR_Flamingos-2_Gagne2015c.fits) FAILED
95/424: 2MASSJ1207-3900_NIR_SpeX_Gagne2014b, chi_r^2 = 19.4, ndof=39
96/424: SIMPJ0135+0205_NIR_SIMON_Robert2016, chi_r^2 = 23.4, ndof=39
97/424: SIMPJ0956-1447_NIR_SpeX_Robert2016, chi_r^2 = 37.4, ndof=39
Unsufficient number of good points (33 < 34). Tmp discarded.
98/424 (2MASSJ2335-1908_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0819-0450_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
99/424 (2MASSJ0819-0450_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Wavelength range of template SIMPJ1158+0435_NIR_SpeX_Robert2016.fits (1.13, 2.43)mu too short compared to that of observed spectrum (0.95, 2.25)mu
100/424 (SIMPJ1158+0435_NIR_SpeX_Robert2016.fits) FAILED
2MASSJ0002+0408A_NIR_SpeX_Gagne2015c.fits could not be opened. Corrupt file?
101/424 (2MASSJ0002+0408A_NIR_SpeX_Gagne2015c.fits) FAILED
Unsufficient number of good points (33 < 34). Tmp discarded.
102/424 (2MASSJ2343-3646_NIR_FIRE_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2335-1908_merge' / [astropy.io.fits.card]
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'W2343-3646_tc.fits' / [astropy.io.fits.card]
103/424: SIMPJ0006-1319_NIR_GNIRS_Robert2016, chi_r^2 = 135.5, ndof=35
104/424: SIMPJ0532-3253_NIR_GNIRS_Robert2016, chi_r^2 = 216.5, ndof=38
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0148-5201_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
105/424 (2MASSJ0148-5201_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ2028-5637_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
106/424 (2MASSJ2028-5637_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2028-5637_ALLCOMB_merge' / [astropy.io.fits.card]
107/424: 2MASSJ0758+15300_NIR_SpeX_Gagne2015c, chi_r^2 = 2.9, ndof=32
108/424: SIMPJ1324+1906_NIR_GNIRS_Robert2016, chi_r^2 = 88.3, ndof=39
Wavelength range of template 2MASSJ2235-5906_NIR_Flamingos-2_Gagne2015c.fits (0.89, 1.89)mu too short compared to that of observed spectrum (0.95, 2.25)mu
109/424 (2MASSJ2235-5906_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0129-0823_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
110/424 (2MASSJ0129-0823_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0819-7401_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
111/424 (2MASSJ0819-7401_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0537-0623_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
112/424 (2MASSJ0537-0623_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2235-5906_BEAMS_AB_140710_386_387_388_389_comb.fits' / [astropy.io.fits.card]
113/424: SIMPJ1307-0558_NIR_GNIRS_Robert2016, chi_r^2 = 262.0, ndof=36
114/424: 2MASSJ1207-3932_NIR_SpeX_Gagne2015c, chi_r^2 = 10.2, ndof=33
Wavelength range of template SIMPJ1118-0856_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
115/424 (SIMPJ1118-0856_NIR_NIRI_Robert2016.fits) FAILED
116/424: SIMPJ1629+0335_NIR_SIMON_Robert2016, chi_r^2 = 49.5, ndof=39
117/424: SIMPJ0357+1529_NIR_GNIRS_Robert2016, chi_r^2 = 20.1, ndof=39
118/424: SIMPJ0400-1322_NIR_GNIRS_Robert2016, chi_r^2 = 124.5, ndof=38
Wavelength range of template 2MASSJ1207-3900_OPT_MAGE_Gagne2014b.fits (38240.71, 43051.20)mu too short compared to that of observed spectrum (0.95, 2.25)mu
119/424 (2MASSJ1207-3900_OPT_MAGE_Gagne2014b.fits) FAILED
120/424: SIMPJ1627+0546_NIR_SpeX_Robert2016, chi_r^2 = 8.2, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2000-7523_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
121/424 (2MASSJ2000-7523_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J20004841-7523070_ALLCOMB_merge' / [astropy.io.fits.card]
WARNING: non-ASCII characters are present in the FITS file header and have been replaced by "?" characters [astropy.io.fits.util]
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'spc0070.a.fits' [astropy.io.fits.card]
122/424: 2MASSJ2154-1055_NIR_SpeX_Gagne2015c, chi_r^2 = 22.7, ndof=39
123/424: 2MASSJ0536-1920_NIR_SpeX_Gagne2015c, chi_r^2 = 39.8, ndof=32
124/424: SIMPJ0422+1033_NIR_SIMON_Robert2016, chi_r^2 = 9.9, ndof=39
Wavelength range of template SIMPJ1333-0215_NIR_SpeX_Robert2016.fits (1.13, 2.43)mu too short compared to that of observed spectrum (0.95, 2.25)mu
125/424 (SIMPJ1333-0215_NIR_SpeX_Robert2016.fits) FAILED
126/424: SIMPJ1452+1114_NIR_SpeX_Robert2016, chi_r^2 = 15.4, ndof=39
Unsufficient number of good points (31 < 34). Tmp discarded.
127/424 (2MASSJ2310-0748_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0649-3823_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
128/424 (2MASSJ0649-3823_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2310-0748_merge' / [astropy.io.fits.card]
129/424: SIMPJ1613-0747_NIR_GNIRS_Robert2016, chi_r^2 = 47.5, ndof=38
130/424: SIMPJ0552+0210_NIR_GNIRS_Robert2016, chi_r^2 = 203.1, ndof=36
131/424: SIMPJ0417-1345_NIR_SpeX_Robert2016, chi_r^2 = 10.5, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0018-6703_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
132/424 (2MASSJ0018-6703_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0300-5459_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
133/424 (2MASSJ0300-5459_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'W2315-4747_tc.fits' / [astropy.io.fits.card]
134/424: 2MASSJ2315-4747_NIR_FIRE_Gagne2015c, chi_r^2 = 13.6, ndof=33
135/424: SIMPJ0316+2650_NIR_SpeX_Robert2016, chi_r^2 = 65.4, ndof=39
Unsufficient number of good points (30 < 34). Tmp discarded.
136/424 (2MASSJ1621-2346_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0320-3313_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
137/424 (2MASSJ0320-3313_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1101-7735_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
138/424 (2MASSJ1101-7735_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Unsufficient number of good points (33 < 34). Tmp discarded.
139/424 (2MASSJ2048-5127_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Wavelength range of template SIMPJ1004-1127_NIR_GNIRS_Robert2016.fits (1.00, 2.52)mu too short compared to that of observed spectrum (0.95, 2.25)mu
140/424 (SIMPJ1004-1127_NIR_GNIRS_Robert2016.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'W2048-5127_tc.fits' / [astropy.io.fits.card]
141/424: 2MASSJ0339-2434_NIR_SpeX_Gagne2015c, chi_r^2 = 3.8, ndof=38
Unsufficient number of good points (33 < 34). Tmp discarded.
142/424 (2MASSJ1529+6312_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0355-1032_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
143/424 (2MASSJ0355-1032_NIR_Flamingos-2_Gagne2015c.fits) FAILED
144/424: SIMPJ0414+1529_NIR_SpeX_Robert2016, chi_r^2 = 7.7, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1257-4111_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
145/424 (2MASSJ1257-4111_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ2048-3255_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
146/424 (2MASSJ2048-3255_NIR_SpeX_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2048-3255_merge' / [astropy.io.fits.card]
147/424: SIMPJ1147+2153_NIR_SpeX_Robert2016, chi_r^2 = 12.5, ndof=39
SIMPJ2332-1249_NIR_SpeX_Robert2016.fits could not be opened. Corrupt file?
148/424 (SIMPJ2332-1249_NIR_SpeX_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ0635-6234_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
149/424 (2MASSJ0635-6234_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (ceil w.r.t 2.36)
Issue with resampling of template SIMPJ0008+0806_NIR_SIMON_Robert2016.fits. Does the wavelength range extend far enough (0.80, 2.35)mu?
150/424 (SIMPJ0008+0806_NIR_SIMON_Robert2016.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2327+3858_merge' / [astropy.io.fits.card]
151/424: 2MASSJ2327+3858_NIR_SpeX_Gagne2015c, chi_r^2 = 4.8, ndof=32
152/424: 2MASSJ1358-0046_NIR_SpeX_Gagne2015c, chi_r^2 = 4.7, ndof=39
153/424: SIMPJ2327+1517_NIR_SpeX_Robert2016, chi_r^2 = 19.8, ndof=39
154/424: SIMPJ0049+0440_NIR_GNIRS_Robert2016, chi_r^2 = 30.8, ndof=39
155/424: 2MASSJ0825-0029_NIR_FIRE_Gagne2015c, chi_r^2 = 9.2, ndof=32
156/424: SIMPJ2154-1055_NIR_SpeX_Gagne2014c, chi_r^2 = 22.7, ndof=39
157/424: SIMPJ0006-1319_NIR_SIMON_Robert2016, chi_r^2 = 27.0, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0244-3548_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
158/424 (2MASSJ0244-3548_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Unsufficient number of good points (29 < 34). Tmp discarded.
159/424 (2MASSJ0342-2904_NIR_FIRE_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ0311+0106_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.46)mu?
160/424 (2MASSJ0311+0106_NIR_Flamingos-2_Gagne2015c.fits) FAILED
161/424: SIMPJ1628+0517_NIR_SIMON_Robert2016, chi_r^2 = 35.5, ndof=39
162/424: SIMPJ1324+1906_NIR_SIMON_Robert2016, chi_r^2 = 77.3, ndof=39
163/424: SIMPJ0921-1534_NIR_GNIRS_Robert2016, chi_r^2 = 105.4, ndof=35
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ1706-1314_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
164/424 (2MASSJ1706-1314_NIR_SpeX_Gagne2015c.fits) FAILED
165/424: SIMPJ1004-1318_NIR_GNIRS_Robert2016, chi_r^2 = 25.8, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0322-7940_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
166/424 (2MASSJ0322-7940_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0051-6227_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
167/424 (2MASSJ0051-6227_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Unsufficient number of good points (31 < 34). Tmp discarded.
168/424 (2MASSJ1642-1942_NIR_FIRE_Gagne2015c.fits) FAILED
169/424: 2MASSJ0953-1014_NIR_SpeX_Gagne2015c, chi_r^2 = 14.0, ndof=39
170/424: SIMPJ1629+0335_NIR_GNIRS_Robert2016, chi_r^2 = 55.5, ndof=37
Wavelength range of template SIMPJ2132-1452_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
171/424 (SIMPJ2132-1452_NIR_NIRI_Robert2016.fits) FAILED
Unsufficient number of good points (32 < 34). Tmp discarded.
172/424 (2MASSJ0825+0340_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2149-6413_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
173/424 (2MASSJ2149-6413_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Wavelength range of template SIMPJ1039-2829_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
174/424 (SIMPJ1039-2829_NIR_NIRI_Robert2016.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2149-6413_ALLCOMB_merge' / [astropy.io.fits.card]
175/424: 2MASSJ0820-7514_NIR_FIRE_Gagne2015c, chi_r^2 = 13.7, ndof=33
176/424: SIMPJ2309+1003_NIR_GNIRS_Robert2016, chi_r^2 = 83.9, ndof=39
177/424: 2MASSJ0805+2505B_NIR_SpeX_Gagne2015c, chi_r^2 = 4.4, ndof=39
178/424: SIMPJ1613-0747_NIR_SIMON_Robert2016, chi_r^2 = 14.0, ndof=39
179/424: 2MASSJ0951+3558_NIR_SpeX_Gagne2015c, chi_r^2 = 3.9, ndof=39
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ2325-0259_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
180/424 (2MASSJ2325-0259_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ0407+1546_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
181/424 (2MASSJ0407+1546_NIR_SpeX_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2325-0259' / [astropy.io.fits.card]
182/424: SIMPJ2254+1640_NIR_SpeX_Robert2016, chi_r^2 = 14.4, ndof=39
183/424: 2MASSJ1623-2353_NIR_SpeX_Gagne2015c, chi_r^2 = 14.9, ndof=39
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template GUPscb_NIR_GNIRS_Naud2014.fits. Does the wavelength range extend far enough (0.94, 2.35)mu?
184/424 (GUPscb_NIR_GNIRS_Naud2014.fits) FAILED
Wavelength range of template SIMPJ1132-3809_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
185/424 (SIMPJ1132-3809_NIR_NIRI_Robert2016.fits) FAILED
186/424: 2MASSJ0526-1824_NIR_SpeX_Gagne2015c, chi_r^2 = 5.0, ndof=34
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1204-2806_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
187/424 (2MASSJ1204-2806_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0540-0923_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
188/424 (2MASSJ0540-0923_NIR_Flamingos-2_Gagne2015c.fits) FAILED
189/424: 2MASSJ0507+1430A_NIR_SpeX_Gagne2015c, chi_r^2 = 3.6, ndof=39
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ2112-8128_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
190/424 (2MASSJ2112-8128_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2112-8128_ALLCOMB_merge' / [astropy.io.fits.card]
191/424: SIMPJ0013+0841_NIR_SIMON_Robert2016, chi_r^2 = 20.7, ndof=39
Unsufficient number of good points (33 < 34). Tmp discarded.
192/424 (2MASSJ1939-5216_NIR_FIRE_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2148-4736_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
193/424 (2MASSJ2148-4736_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'W1939-5216_tc.fits' / [astropy.io.fits.card]
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2148-4736_ALLCOMB_merge' / [astropy.io.fits.card]
194/424: SIMPJ1117+1857_NIR_GNIRS_Robert2016, chi_r^2 = 286.3, ndof=36
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ0355+1133_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
195/424 (2MASSJ0355+1133_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2005-6258_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
196/424 (2MASSJ2005-6258_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ0211-1427_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
197/424 (SIMPJ0211-1427_NIR_SpeX_Robert2016.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2005-6258_ALLCOMB_merge' / [astropy.io.fits.card]
198/424: SIMPJ1041-0429_NIR_GNIRS_Robert2016, chi_r^2 = 21.0, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0804-6346_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
199/424 (2MASSJ0804-6346_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Wavelength range of template SIMPJ1343-1216_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
200/424 (SIMPJ1343-1216_NIR_NIRI_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1903-3723_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
201/424 (2MASSJ1903-3723_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J1903-3723_ALLCOMB_merge' / [astropy.io.fits.card]
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J1948+5944A_merge' / [astropy.io.fits.card]
202/424: 2MASSJ1948+5944A_NIR_SpeX_Gagne2015c, chi_r^2 = 3.1, ndof=32
Wavelength range of template 2MASSJ1846-5706_NIR_Flamingos-2_Gagne2015c.fits (0.90, 1.90)mu too short compared to that of observed spectrum (0.95, 2.25)mu
203/424 (2MASSJ1846-5706_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ0453-1751_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
204/424 (2MASSJ0453-1751_NIR_SpeX_Gagne2015c.fits) FAILED
Unsufficient number of good points (33 < 34). Tmp discarded.
205/424 (2MASSJ2048-5127_NIR_FIRE_Gagne2015c.fits) FAILED
206/424: SIMPJ1537-0800_NIR_GNIRS_Robert2016, chi_r^2 = 169.0, ndof=36
Wavelength range of template SIMPJ2322-1407_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
207/424 (SIMPJ2322-1407_NIR_NIRI_Robert2016.fits) FAILED
208/424: SIMPJ2132-1452_NIR_SIMON_Robert2016, chi_r^2 = 102.1, ndof=39
209/424: 2MASSJ0404+2616A_NIR_SpeX_Gagne2015c, chi_r^2 = 6.8, ndof=33
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ1547-1626A_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
210/424 (2MASSJ1547-1626A_NIR_SpeX_Gagne2015c.fits) FAILED
211/424: SIMPJ0425-1900_NIR_GNIRS_Robert2016, chi_r^2 = 37.9, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1249-2035_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
212/424 (2MASSJ1249-2035_NIR_Flamingos-2_Gagne2015c.fits) FAILED
213/424: SIMPJ0120+1518_NIR_SpeX_Robert2016, chi_r^2 = 11.2, ndof=39
214/424: SIMPJ2239+1617_NIR_SIMON_Robert2016, chi_r^2 = 61.1, ndof=39
215/424: SIMPJ1708+2606_NIR_SpeX_Robert2016, chi_r^2 = 24.5, ndof=39
216/424: SIMPJ1036+0306_NIR_SIMON_Robert2016, chi_r^2 = 23.2, ndof=39
217/424: SIMPJ0421-1133_NIR_GNIRS_Robert2016, chi_r^2 = 189.6, ndof=35
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1221+0257_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
218/424 (2MASSJ1221+0257_NIR_Flamingos-2_Gagne2015c.fits) FAILED
219/424: SIMPJ1014+1900_NIR_GNIRS_Robert2016, chi_r^2 = 52.9, ndof=36
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0910-7552_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
220/424 (2MASSJ0910-7552_NIR_Flamingos-2_Gagne2015c.fits) FAILED
221/424: 2MASSJ0520+0511_NIR_SpeX_Gagne2015c, chi_r^2 = 6.9, ndof=32
222/424: SIMPJ0936+0528_NIR_GNIRS_Robert2016, chi_r^2 = 224.4, ndof=38
223/424: SIMPJ1052+1722_NIR_SIMON_Robert2016, chi_r^2 = 14.6, ndof=39
224/424: SIMPJ2117-0611_NIR_SIMON_Robert2016, chi_r^2 = 19.9, ndof=39
No indices match the constraint (floor w.r.t 0.84)
Issue with resampling of template 2MASSJ1625-2358_NIR_GNIRS_Gagne2015c.fits. Does the wavelength range extend far enough (0.84, 2.53)mu?
225/424 (2MASSJ1625-2358_NIR_GNIRS_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_F-TLM= '2013-07-18T21:19:33' / Time of last modification [astropy.io.fits.card]
226/424: SIMPJ0251-0818_NIR_SIMON_Robert2016, chi_r^2 = 17.0, ndof=39
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J1846+5246_A_merge' / [astropy.io.fits.card]
227/424: 2MASSJ1846+5246A_NIR_SpeX_Gagne2015c, chi_r^2 = 8.0, ndof=32
2MASSJ0039+1330A_NIR_SpeX_Gagne2015c.fits could not be opened. Corrupt file?
228/424 (2MASSJ0039+1330A_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ1257-3635_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
229/424 (2MASSJ1257-3635_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J12574463-3635431_ALLCOMB_merge' / [astropy.io.fits.card]
230/424: SIMPJ1150+0520_NIR_SpeX_Robert2016, chi_r^2 = 20.2, ndof=39
231/424: 2MASSJ1013-1706B_NIR_SpeX_Gagne2015c, chi_r^2 = 3.1, ndof=39
232/424: SIMPJ1141+1941_NIR_GNIRS_Robert2016, chi_r^2 = 22.8, ndof=35
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ1003-1441_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.43)mu?
233/424 (SIMPJ1003-1441_NIR_SpeX_Robert2016.fits) FAILED
234/424: SIMPJ1631-1922_NIR_SpeX_Robert2016, chi_r^2 = 3.5, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2235-3844_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
235/424 (2MASSJ2235-3844_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ0326-0617_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
236/424 (2MASSJ0326-0617_NIR_SpeX_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2235-3844_ALLCOMB_merge' / [astropy.io.fits.card]
237/424: 2MASSJ0518-3101_NIR_SpeX_Gagne2015c, chi_r^2 = 7.2, ndof=37
238/424: SIMPJ1422+0827_NIR_SIMON_Robert2016, chi_r^2 = 31.3, ndof=39
2MASSJ2331-0406_NIR_Flamingos-2_Gagne2015c.fits could not be opened. Corrupt file?
239/424 (2MASSJ2331-0406_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: VerifyWarning: Error validating header for HDU #0 (note: Astropy uses zero-based indexing).
Header size is not multiple of 2880: 11366
There may be extra bytes after the last HDU or the file is corrupted. [astropy.io.fits.hdu.hdulist]
240/424: SIMPJ1339+1523_NIR_SIMON_Robert2016, chi_r^2 = 13.0, ndof=39
241/424: 2MASSJ1226-3316_NIR_SpeX_Gagne2015c, chi_r^2 = 3.4, ndof=32
242/424: SIMPJ1615+1340_NIR_SIMON_Robert2016, chi_r^2 = 135.2, ndof=39
243/424: 2MASSJ0758+15301B_NIR_SpeX_Gagne2015c, chi_r^2 = 3.6, ndof=32
244/424: 2MASSJ1247-3816_NIR_SpeX_Gagne2014b, chi_r^2 = 9.2, ndof=39
245/424: SIMPJ0026-0936_NIR_SpeX_Robert2016, chi_r^2 = 8.3, ndof=39
246/424: 2MASSJ1005+1703_NIR_SpeX_Gagne2015c, chi_r^2 = 4.2, ndof=39
2MASSJ0017-3219_NIR_Flamingos-2_Gagne2015c.fits could not be opened. Corrupt file?
247/424 (2MASSJ0017-3219_NIR_Flamingos-2_Gagne2015c.fits) FAILED
248/424: SIMPJ1318-0632_NIR_SIMON_Robert2016, chi_r^2 = 15.1, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0945-7753_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
249/424 (2MASSJ0945-7753_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0240-4253_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
250/424 (2MASSJ0240-4253_NIR_Flamingos-2_Gagne2015c.fits) FAILED
251/424: SIMPJ2257-0140_NIR_SIMON_Robert2016, chi_r^2 = 96.7, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0126-5505_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
252/424 (2MASSJ0126-5505_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1510-2818_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
253/424 (2MASSJ1510-2818_NIR_Flamingos-2_Gagne2015c.fits) FAILED
254/424: 2MASSJ1148-2836_NIR_SpeX_Gagne2015c, chi_r^2 = 15.1, ndof=39
255/424: 2MASSJ1028-2830_NIR_SpeX_Gagne2015c, chi_r^2 = 3.2, ndof=33
256/424: SIMPJ1344-1614_NIR_SpeX_Robert2016, chi_r^2 = 39.5, ndof=39
257/424: SIMPJ1122+0343_NIR_SIMON_Robert2016, chi_r^2 = 13.4, ndof=39
258/424: SIMPJ0103+1940_NIR_SpeX_Robert2016, chi_r^2 = 8.0, ndof=39
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ0417+1634_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
259/424 (SIMPJ0417+1634_NIR_SpeX_Robert2016.fits) FAILED
260/424: SIMPJ1308+0432_NIR_SIMON_Robert2016, chi_r^2 = 15.7, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1935-6200_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
261/424 (2MASSJ1935-6200_NIR_Flamingos-2_Gagne2015c.fits) FAILED
SIMPJ2344+0909_NIR_GNIRS_Robert2016.fits could not be opened. Corrupt file?
262/424 (SIMPJ2344+0909_NIR_GNIRS_Robert2016.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J19350976-6200473_ALLCOMB_merge' / [astropy.io.fits.card]
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2202-1109_merge' / [astropy.io.fits.card]
263/424: 2MASSJ2202-1109_NIR_SpeX_Gagne2015c, chi_r^2 = 5.5, ndof=32
2MASSJ0103-2805A_NIR_SpeX_Gagne2015c.fits could not be opened. Corrupt file?
264/424 (2MASSJ0103-2805A_NIR_SpeX_Gagne2015c.fits) FAILED
Unsufficient number of good points (33 < 34). Tmp discarded.
265/424 (2MASSJ0019+4614_NIR_SpeX_Gagne2015c.fits) FAILED
266/424: SIMPJ1537-0800_NIR_SIMON_Robert2016, chi_r^2 = 19.1, ndof=39
267/424: SIMPJ0858+2214_NIR_SIMON_Robert2016, chi_r^2 = 15.3, ndof=39
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ1654+3747_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.43)mu?
268/424 (SIMPJ1654+3747_NIR_SpeX_Robert2016.fits) FAILED
269/424: SIMPJ1044+0620_NIR_SIMON_Robert2016, chi_r^2 = 12.4, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0210-3015_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
270/424 (2MASSJ0210-3015_NIR_Flamingos-2_Gagne2015c.fits) FAILED
271/424: SIMPJ2203-0301_NIR_SpeX_Robert2016, chi_r^2 = 10.6, ndof=39
272/424: SIMPJ0946+0922_NIR_GNIRS_Robert2016, chi_r^2 = 272.7, ndof=36
273/424: SIMPJ0307+0852_NIR_GNIRS_Robert2016, chi_r^2 = 207.5, ndof=38
Wavelength range of template 2MASSJ2336-3541_NIR_Flamingos-2_Gagne2015c.fits (0.90, 1.89)mu too short compared to that of observed spectrum (0.95, 2.25)mu
274/424 (2MASSJ2336-3541_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2336-3541_BEAMS_AB_140710_397_398_399_400_comb.fits' / [astropy.io.fits.card]
275/424: SIMPJ1036+0306_NIR_GNIRS_Robert2016, chi_r^2 = 63.0, ndof=38
No indices match the constraint (floor w.r.t 0.91)
Issue with resampling of template 2MASSJ2033-3733_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.91, 2.46)mu?
276/424 (2MASSJ2033-3733_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Unsufficient number of good points (33 < 34). Tmp discarded.
277/424 (2MASSJ1256-2718_NIR_FIRE_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2033-3733_ALLCOMB_merge' / [astropy.io.fits.card]
278/424: SIMPJ1052+1722_NIR_GNIRS_Robert2016, chi_r^2 = 230.3, ndof=38
279/424: SIMPJ1605+1931_NIR_SpeX_Robert2016, chi_r^2 = 16.3, ndof=39
280/424: 2MASSJ0058-0651_NIR_SpeX_Gagne2015c, chi_r^2 = 11.5, ndof=32
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0708-4701_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
281/424 (2MASSJ0708-4701_NIR_Flamingos-2_Gagne2015c.fits) FAILED
282/424: SIMPJ0423+1212_NIR_GNIRS_Robert2016, chi_r^2 = 16.1, ndof=39
283/424: SIMPJ0422+0723_NIR_SpeX_Robert2016, chi_r^2 = 13.9, ndof=39
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ2033-5635_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
284/424 (2MASSJ2033-5635_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Unsufficient number of good points (32 < 34). Tmp discarded.
285/424 (2MASSJ2114-4339_NIR_SpeX_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2114-4339_merge' / [astropy.io.fits.card]
286/424: SIMPJ0915+0514_NIR_SpeX_Robert2016, chi_r^2 = 14.1, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2050-3639_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
287/424 (2MASSJ2050-3639_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J20505221-3639552_ALLCOMB_merge' / [astropy.io.fits.card]
288/424: SIMPJ2304+0749_NIR_GNIRS_Robert2016, chi_r^2 = 16.5, ndof=36
289/424: SIMPJ0136+0933_NIR_GNIRS_Robert2016, chi_r^2 = 79.7, ndof=36
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ0034-4102_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
290/424 (2MASSJ0034-4102_NIR_Flamingos-2_Gagne2015c.fits) FAILED
291/424: SIMPJ1627+0546_NIR_SIMON_Robert2016, chi_r^2 = 12.1, ndof=39
292/424: SIMPJ0013-1816_NIR_SpeX_Robert2016, chi_r^2 = 23.4, ndof=39
293/424: SIMPJ2330+1006_NIR_GNIRS_Robert2016, chi_r^2 = 297.0, ndof=38
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2322-6151A_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
294/424 (2MASSJ2322-6151A_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2322-6151_ALLCOMB_merge' / [astropy.io.fits.card]
295/424: SIMPJ1138+0748_NIR_SIMON_Robert2016, chi_r^2 = 48.5, ndof=39
296/424: 2MASSJ0548-2942_NIR_SpeX_Gagne2015c, chi_r^2 = 6.6, ndof=36
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ1622-2346_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
297/424 (2MASSJ1622-2346_NIR_SpeX_Gagne2015c.fits) FAILED
298/424: SIMPJ1343+1312_NIR_GNIRS_Robert2016, chi_r^2 = 14.8, ndof=39
Wavelength range of template SIMPJ0006-2158_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
299/424 (SIMPJ0006-2158_NIR_NIRI_Robert2016.fits) FAILED
Unsufficient number of good points (33 < 34). Tmp discarded.
300/424 (2MASSJ2353-1844B_NIR_SpeX_Gagne2015c.fits) FAILED
Wavelength range of template SIMPJ1615+1340_NIR_GNIRS_Robert2016.fits (1.00, 2.52)mu too short compared to that of observed spectrum (0.95, 2.25)mu
301/424 (SIMPJ1615+1340_NIR_GNIRS_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ0501-0010_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
302/424 (2MASSJ0501-0010_NIR_SpeX_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2329_tell.fits' / [astropy.io.fits.card]
303/424: 2MASSJ2329+0329_NIR_SpeX_Gagne2015c, chi_r^2 = 7.9, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0540-0906_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
304/424 (2MASSJ0540-0906_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Wavelength range of template DENISJ0817-6155_NIR_OSIRIS_Artigau2010.fits (1.13, 2.36)mu too short compared to that of observed spectrum (0.95, 2.25)mu
305/424 (DENISJ0817-6155_NIR_OSIRIS_Artigau2010.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0228+0218_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
306/424 (2MASSJ0228+0218_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ1051-1916_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
307/424 (2MASSJ1051-1916_NIR_SpeX_Gagne2015c.fits) FAILED
SIMPJ2351+3010_NIR_SpeX_Robert2016.fits could not be opened. Corrupt file?
308/424 (SIMPJ2351+3010_NIR_SpeX_Robert2016.fits) FAILED
309/424: SIMPJ1339+1523_NIR_GNIRS_Robert2016, chi_r^2 = 283.0, ndof=37
310/424: SIMPJ0812+3721_NIR_SpeX_Robert2016, chi_r^2 = 9.6, ndof=39
311/424: SIMPJ1422+0827_NIR_GNIRS_Robert2016, chi_r^2 = 122.3, ndof=38
312/424: SIMPJ2143-1544_NIR_SpeX_Robert2016, chi_r^2 = 9.1, ndof=39
313/424: SIMPJ2245+1722_NIR_SIMON_Robert2016, chi_r^2 = 19.9, ndof=39
314/424: GUPscA_NIR_SpeX_Naud2014, chi_r^2 = 3.3, ndof=39
Unsufficient number of good points (33 < 34). Tmp discarded.
315/424 (2MASSJ1839+2952_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ2002-1316_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
316/424 (2MASSJ2002-1316_NIR_SpeX_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J1839+2952_merge' / [astropy.io.fits.card]
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2002-1316_merge' / [astropy.io.fits.card]
317/424: SIMPJ1147-1032_NIR_GNIRS_Robert2016, chi_r^2 = 226.5, ndof=39
Wavelength range of template SIMPJ1039+2440_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
318/424 (SIMPJ1039+2440_NIR_NIRI_Robert2016.fits) FAILED
319/424: 2MASSJ0344+0716A_NIR_SpeX_Gagne2015c, chi_r^2 = 3.1, ndof=33
Wavelength range of template SIMPJ0004-2007_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
320/424 (SIMPJ0004-2007_NIR_NIRI_Robert2016.fits) FAILED
321/424: 2MASSJ0336-2619_NIR_SpeX_Gagne2015c, chi_r^2 = 4.8, ndof=39
322/424: SIMPJ0946+1808_NIR_SIMON_Robert2016, chi_r^2 = 11.3, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0545-0121_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
323/424 (2MASSJ0545-0121_NIR_Flamingos-2_Gagne2015c.fits) FAILED
324/424: SIMPJ1003-1441_NIR_SIMON_Robert2016, chi_r^2 = 7.4, ndof=39
325/424: SIMPJ1308+0432_NIR_GNIRS_Robert2016, chi_r^2 = 290.2, ndof=37
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ1623-0508_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.43)mu?
326/424 (SIMPJ1623-0508_NIR_SpeX_Robert2016.fits) FAILED
327/424: CFBDSIR2149_OPT+NIR_X-Shooter_Delorme2012, chi_r^2 = 166.3, ndof=39
Wavelength range of template SIMPJ1533+2044_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
328/424 (SIMPJ1533+2044_NIR_NIRI_Robert2016.fits) FAILED
329/424: SIMPJ1200-2836_NIR_SpeX_Robert2016, chi_r^2 = 30.8, ndof=39
330/424: SIMPJ2250+0808_NIR_SpeX_Robert2016, chi_r^2 = 28.4, ndof=39
331/424: SIMPJ1510-1147_NIR_SIMON_Robert2016, chi_r^2 = 19.9, ndof=39
332/424: 2MASSJ0814+0253_NIR_SpeX_Gagne2015c, chi_r^2 = 3.4, ndof=32
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ1200-3405_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
333/424 (2MASSJ1200-3405_NIR_Flamingos-2_Gagne2015c.fits) FAILED
334/424: SIMPJ1035+3655_NIR_SpeX_Robert2016, chi_r^2 = 7.8, ndof=39
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2039-1126_merge' / [astropy.io.fits.card]
335/424: 2MASSJ2039-1126_NIR_SpeX_Gagne2015c, chi_r^2 = 6.6, ndof=35
336/424: 2MASSJ0803+0827_NIR_SpeX_Gagne2015c, chi_r^2 = 3.8, ndof=33
337/424: SIMPJ2157-0130_NIR_SIMON_Robert2016, chi_r^2 = 19.1, ndof=39
338/424: 2MASSJ0805+2505A_NIR_SpeX_Gagne2015c, chi_r^2 = 3.8, ndof=39
339/424: SIMPJ1256-1002_NIR_SpeX_Robert2016, chi_r^2 = 7.7, ndof=39
340/424: SIMPJ1118-0856_NIR_SpeX_Robert2016, chi_r^2 = 26.9, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2313-6127_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
341/424 (2MASSJ2313-6127_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2313-6127_ALLCOMB_merge' / [astropy.io.fits.card]
342/424: SIMPJ0410+1459_NIR_SpeX_Robert2016, chi_r^2 = 12.0, ndof=39
343/424: SIMPJ1009+1559_NIR_GNIRS_Robert2016, chi_r^2 = 254.7, ndof=35
344/424: SIMPJ2213+1255_NIR_SpeX_Robert2016, chi_r^2 = 7.0, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0027-0806_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
345/424 (2MASSJ0027-0806_NIR_Flamingos-2_Gagne2015c.fits) FAILED
346/424: 2MASSJ0507+1430B_NIR_SpeX_Gagne2015c, chi_r^2 = 3.3, ndof=39
347/424: SIMPJ0138-0322_NIR_SIMON_Robert2016, chi_r^2 = 64.2, ndof=39
348/424: SIMPJ0155+0950_NIR_SIMON_Robert2016, chi_r^2 = 18.1, ndof=39
No indices match the constraint (floor w.r.t 0.85)
Issue with resampling of template 2MASSJ1252-3415_NIR_GNIRS_Gagne2015c.fits. Does the wavelength range extend far enough (0.85, 2.52)mu?
349/424 (2MASSJ1252-3415_NIR_GNIRS_Gagne2015c.fits) FAILED
350/424: 2MASSJ0153-6744_NIR_FIRE_Gagne2015c, chi_r^2 = 12.5, ndof=32
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2322-6151B_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
351/424 (2MASSJ2322-6151B_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2322-6151B_ALLCOMB_merge' / [astropy.io.fits.card]
352/424: SIMPJ1811+2728_NIR_SpeX_Robert2016, chi_r^2 = 21.2, ndof=39
No indices match the constraint (floor w.r.t 0.84)
Issue with resampling of template 2MASSJ1627-2411_NIR_GNIRS_Gagne2015c.fits. Does the wavelength range extend far enough (0.84, 2.53)mu?
353/424 (2MASSJ1627-2411_NIR_GNIRS_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_F-TLM= '2013-07-18T20:09:23' / Time of last modification [astropy.io.fits.card]
354/424: 2MASSJ0021-4244_NIR_SpeX_Gagne2015c, chi_r^2 = 13.1, ndof=34
Unsufficient number of good points (33 < 34). Tmp discarded.
355/424 (2MASSJ2206-6116_NIR_FIRE_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'W2206-6116_tc.fits' / [astropy.io.fits.card]
356/424: SIMPJ2255-0725_NIR_SIMON_Robert2016, chi_r^2 = 38.5, ndof=39
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ1455+2619_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.43)mu?
357/424 (SIMPJ1455+2619_NIR_SpeX_Robert2016.fits) FAILED
Unsufficient number of good points (29 < 34). Tmp discarded.
358/424 (2MASSJ0258-1520_NIR_FIRE_Gagne2015c.fits) FAILED
359/424: SIMPJ1127+1234_NIR_GNIRS_Robert2016, chi_r^2 = 117.2, ndof=36
Wavelength range of template SIMPJ1756+3343_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
360/424 (SIMPJ1756+3343_NIR_NIRI_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.85)
Issue with resampling of template SIMPJ0358+1039_NIR_SIMON_Robert2016.fits. Does the wavelength range extend far enough (0.85, 2.44)mu?
361/424 (SIMPJ0358+1039_NIR_SIMON_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2257-5041_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
362/424 (2MASSJ2257-5041_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template 2MASSJ1301-1510_NIR_SpeX_Gagne2015c.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
363/424 (2MASSJ1301-1510_NIR_SpeX_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2257-5041_ALLCOMB_merge' / [astropy.io.fits.card]
364/424: SIMPJ1130+2341_NIR_SpeX_Robert2016, chi_r^2 = 25.9, ndof=39
365/424: SIMPJ0357-1937_NIR_SpeX_Robert2016, chi_r^2 = 8.9, ndof=39
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ2154-7459_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
366/424 (2MASSJ2154-7459_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2154-7459_ALLCOMB_merge' / [astropy.io.fits.card]
367/424: 2MASSJ2206-4217_NIR_SpeX_Gagne2015c, chi_r^2 = 17.3, ndof=39
Wavelength range of template SIMPJ1143+1905_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
368/424 (SIMPJ1143+1905_NIR_NIRI_Robert2016.fits) FAILED
Wavelength range of template SIMPJ2315+2235_NIR_NIRI_Robert2016.fits (0.98, 2.45)mu too short compared to that of observed spectrum (0.95, 2.25)mu
369/424 (SIMPJ2315+2235_NIR_NIRI_Robert2016.fits) FAILED
370/424: SIMPJ2248-0126_NIR_SpeX_Robert2016, chi_r^2 = 17.6, ndof=39
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ1542-3358_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.46)mu?
371/424 (2MASSJ1542-3358_NIR_Flamingos-2_Gagne2015c.fits) FAILED
372/424: SIMPJ0432-0639_NIR_SpeX_Robert2016, chi_r^2 = 11.7, ndof=39
373/424: SIMPJ1644+2600_NIR_SpeX_Robert2016, chi_r^2 = 16.3, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0006-6436_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
374/424 (2MASSJ0006-6436_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0253-7959_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
375/424 (2MASSJ0253-7959_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ0241-5511_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
376/424 (2MASSJ0241-5511_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Wavelength range of template SIMPJ0102+0355_NIR_GNIRS_Robert2016.fits (1.00, 2.52)mu too short compared to that of observed spectrum (0.95, 2.25)mu
377/424 (SIMPJ0102+0355_NIR_GNIRS_Robert2016.fits) FAILED
378/424: 2MASSJ0449+1607_NIR_SpeX_Gagne2015c, chi_r^2 = 15.5, ndof=39
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ0940+2946_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
379/424 (SIMPJ0940+2946_NIR_SpeX_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0512-3041_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
380/424 (2MASSJ0512-3041_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0250-0151_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
381/424 (2MASSJ0250-0151_NIR_Flamingos-2_Gagne2015c.fits) FAILED
382/424: SIMPJ0155+0950_NIR_GNIRS_Robert2016, chi_r^2 = 26.4, ndof=39
Unsufficient number of good points (33 < 34). Tmp discarded.
383/424 (2MASSJ0046+0252_NIR_SpeX_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ0956-1910_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.43)mu?
384/424 (SIMPJ0956-1910_NIR_SpeX_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2038-4118_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
385/424 (2MASSJ2038-4118_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2038-4118_ALLCOMB_merge' / [astropy.io.fits.card]
386/424: SIMPJ1013-1946_NIR_GNIRS_Robert2016, chi_r^2 = 270.5, ndof=37
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2314-5405_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
387/424 (2MASSJ2314-5405_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0440-1820_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
388/424 (2MASSJ0440-1820_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2314-5405_ALLCOMB_merge' / [astropy.io.fits.card]
389/424: SIMPJ1108+0838_NIR_GNIRS_Robert2016, chi_r^2 = 43.2, ndof=36
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0004-1709_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
390/424 (2MASSJ0004-1709_NIR_Flamingos-2_Gagne2015c.fits) FAILED
391/424: 2MASSJ1325+0600_NIR_SpeX_Gagne2015c, chi_r^2 = 11.4, ndof=39
392/424: SIMPJ2235+0418_NIR_SIMON_Robert2016, chi_r^2 = 18.9, ndof=39
2MASSJ0017-0316_NIR_SpeX_Gagne2015c.fits could not be opened. Corrupt file?
393/424 (2MASSJ0017-0316_NIR_SpeX_Gagne2015c.fits) FAILED
394/424: SIMPJ0421-1950_NIR_GNIRS_Robert2016, chi_r^2 = 195.8, ndof=35
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0417-1140_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
395/424 (2MASSJ0417-1140_NIR_Flamingos-2_Gagne2015c.fits) FAILED
396/424: 2MASSJ0541-0737_NIR_SpeX_Gagne2015c, chi_r^2 = 12.9, ndof=32
No indices match the constraint (floor w.r.t 0.92)
Issue with resampling of template 2MASSJ2134-1840_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.92, 2.46)mu?
397/424 (2MASSJ2134-1840_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Wavelength range of template 2MASSJ1253-4211_NIR_Flamingos-2_Gagne2015c.fits (0.90, 1.89)mu too short compared to that of observed spectrum (0.95, 2.25)mu
398/424 (2MASSJ1253-4211_NIR_Flamingos-2_Gagne2015c.fits) FAILED
Unsufficient number of good points (29 < 34). Tmp discarded.
399/424 (2MASSJ1633-2425_NIR_SpeX_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2134-1840_ALLCOMB_merge' / [astropy.io.fits.card]
400/424: SIMPJ1102+1629_NIR_GNIRS_Robert2016, chi_r^2 = 73.6, ndof=36
401/424: SIMPJ0945-0757_NIR_SpeX_Robert2016, chi_r^2 = 17.4, ndof=39
402/424: SIMPJ2322+1300_NIR_SIMON_Robert2016, chi_r^2 = 18.7, ndof=39
No indices match the constraint (floor w.r.t 0.90)
Issue with resampling of template 2MASSJ0318-3708_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.90, 2.47)mu?
403/424 (2MASSJ0318-3708_NIR_Flamingos-2_Gagne2015c.fits) FAILED
404/424: SIMPJ1343-1216_NIR_SpeX_Robert2016, chi_r^2 = 35.3, ndof=39
405/424: 2MASSJ0527+0007A_NIR_SpeX_Gagne2015c, chi_r^2 = 7.3, ndof=39
406/424: SIMPJ1218-1332_NIR_GNIRS_Robert2016, chi_r^2 = 293.4, ndof=36
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0854-3051_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
407/424 (2MASSJ0854-3051_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0720-5617_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
408/424 (2MASSJ0720-5617_NIR_Flamingos-2_Gagne2015c.fits) FAILED
409/424: SIMPJ0006-0852_NIR_GNIRS_Robert2016, chi_r^2 = 19.8, ndof=39
410/424: SIMPJ0331+1944_NIR_SIMON_Robert2016, chi_r^2 = 29.3, ndof=39
2MASSJ0002+0408B_NIR_SpeX_Gagne2015c.fits could not be opened. Corrupt file?
411/424 (2MASSJ0002+0408B_NIR_SpeX_Gagne2015c.fits) FAILED
412/424: 2MASSJ0524+0640_NIR_SpeX_Gagne2015c, chi_r^2 = 4.2, ndof=35
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0440-5126_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
413/424 (2MASSJ0440-5126_NIR_Flamingos-2_Gagne2015c.fits) FAILED
414/424: 2MASSJ0720-0846_NIR_SpeX_Gagne2015c, chi_r^2 = 13.3, ndof=39
415/424: SIMPJ1118-0856_NIR_SIMON_Robert2016, chi_r^2 = 17.8, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0134-5707_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
416/424 (2MASSJ0134-5707_NIR_Flamingos-2_Gagne2015c.fits) FAILED
417/424: SIMPJ2233-1331_NIR_SIMON_Robert2016, chi_r^2 = 43.7, ndof=39
418/424: 2MASSJ0333-3215_NIR_SpeX_Gagne2015c, chi_r^2 = 4.1, ndof=33
419/424: SIMPJ0430+1035_NIR_GNIRS_Robert2016, chi_r^2 = 254.0, ndof=39
No indices match the constraint (floor w.r.t 0.94)
Issue with resampling of template SIMPJ0151+3824_NIR_SpeX_Robert2016.fits. Does the wavelength range extend far enough (0.94, 2.42)mu?
420/424 (SIMPJ0151+3824_NIR_SpeX_Robert2016.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ2022-5645_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.46)mu?
421/424 (2MASSJ2022-5645_NIR_Flamingos-2_Gagne2015c.fits) FAILED
WARNING: The following header keyword is invalid or follows an unrecognized non-standard convention:
I_R_A_FNAME= 'J2022-5645_ALLCOMB_merge' / [astropy.io.fits.card]
422/424: 2MASSJ1247-3816_NIR_SpeX_Gagne2015c, chi_r^2 = 9.2, ndof=39
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0418-4507_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
423/424 (2MASSJ0418-4507_NIR_Flamingos-2_Gagne2015c.fits) FAILED
No indices match the constraint (floor w.r.t 0.89)
Issue with resampling of template 2MASSJ0423-1533_NIR_Flamingos-2_Gagne2015c.fits. Does the wavelength range extend far enough (0.89, 2.47)mu?
424/424 (2MASSJ0423-1533_NIR_Flamingos-2_Gagne2015c.fits) FAILED
****************************************
228/424 template spectra were fitted.
Running time: 0:08:56.400537
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
best chi: [2.6494487 2.71261062 2.87087266]
best scale fac: [8.93336943e-05 5.61832126e-05 9.14677818e-03]
n_dof: [33. 33. 32.]
The best template #1 is: 2MASSJ1119-3917_NIR_SpeX_Gagne2015c.fits (Delta A_V=2.4mag)
The best template #2 is: 2MASSJ1153-3015_NIR_SpeX_Gagne2015c.fits (Delta A_V=2.4mag)
The best template #3 is: 2MASSJ0758+15300_NIR_SpeX_Gagne2015c.fits (Delta A_V=2.2mag)
Let’s load the results, and prepare a list of short template names (for display; i.e. without the suffix):
[141]:
final_tmpname, final_tmp, final_chi, final_params, final_ndof = final_res
n_end = len('_NIR_SpeX_Gagne2015c.fits')
short_tmpname = [final_tmpname[i][:-n_end] for i in range(len(final_tmpname))]
short_tmpname
[141]:
['2MASSJ1119-3917', '2MASSJ1153-3015', '2MASSJ0758+15300']
Checking in the online table of the MSL, these spectra turn out to correspond to the following types of objects:
[142]:
labels_tmp = ["M3", "M4.5", "M4"]
Feel free to repeat the fit after changing the exact value of min_npts (e.g.~lower this value to allow more templates to be considered in the search). Although the best-fit templates may change a little, you should mostly retrieve M3-M5 spectral types as top-3 template spectra for reasonable values of min_npts.
Let’s finally plot the best-fit template spectra, at native resolution and resampled, respectively:
[143]:
from special.model_resampling import resample_model
# Prepare plot
fig, (ax1, ax2) = plt.subplots(2,1,figsize=(11,12))
ax1.set_xlabel(r"Wavelength ($\mu$m)")
ax1.set_ylabel("$\lambda F_{\lambda}$ (W m$^{-2}$)")
ax1.set_ylim(0.8*np.amin(lbda*(spec+spec_err)),
1.1*np.amax(lbda*(spec+spec_err)))
ax2.set_xlabel(r"Wavelength ($\mu$m)")
ax2.set_ylabel("$\lambda F_{\lambda}$ (W m$^{-2}$)")
ax2.set_ylim(0.8*np.amin(lbda*(spec+spec_err)),
1.1*np.amax(lbda*(spec+spec_err)))
# plot options
cols = ['k', 'r', 'b', 'c', 'y', 'm'] # colors of different models
maj_tick_sp = 0.5 # WL spacing for major ticks
min_tick_sp = maj_tick_sp / 5. # WL spacing for minor ticks
# Plot of top 3 spectra:
counter = 0
object_list = []
for ll in range(n_best):
## loading
try:
_, header = open_fits(inpath_models+final_tmpname[ll],header=True)
object_list.append(header['OBJECT'])
except KeyError:
object_list.append(short_tmpname[ll])
lbda_tmp, flux_tmp, flux_tmp_err = final_tmp[ll]
## cropping range
try:
idx_ini = find_nearest(lbda_tmp,0.98*lbda_crop[0], constraint='floor')
except:
idx_ini = 0
try:
idx_fin = find_nearest(lbda_tmp,1.02*lbda_crop[-1], constraint='ceil')
except:
idx_fin = -1
lbda_tmp_crop = lbda_tmp[idx_ini:idx_fin]
flux_tmp_crop = flux_tmp[idx_ini:idx_fin]
dlbda_tmp = np.mean(lbda_tmp_crop[1:]-lbda_tmp_crop[:-1])
## Convolving + resampling template spectrum
tmp_res = resample_model(lbda_crop, lbda_tmp, flux_tmp,
dlbda_obs=dlbda_crop,
instru_res=instru_res_crop,
instru_idx=instru_idx_crop,
filter_reader=filter_reader)
lbda_tmp_res, flux_tmp_res = tmp_res
## formatting labels
lab_str_list = [labels_tmp[ll]]
if AV_range is not None:
lab_str = r'$\Delta A_V$={0:.{1}f}{2}'.format(final_params[1][ll],1,'mag')
lab_str_list.append(lab_str)
sep = '; '
if n_best>1:
label = "Template #{:.0f}: {} ({})".format(ll+1, short_tmpname[ll], #labels_tmp[ll],
sep.join(lab_str_list))
else:
label = "Best template: {} ({})".format(short_tmpname[ll], #labels_tmp[ll],
sep.join(lab_str_list))
## plotting
ax1.plot(lbda_tmp_crop, lbda_tmp_crop*flux_tmp_crop, cols[ll+1],
alpha=0.6, label=label)
ax2.plot(lbda_tmp_res, lbda_tmp_res*flux_tmp_res, cols[ll+1],
alpha=0.6, label=label)
## ticks
min_tick = lbda_crop[0]-dlbda_crop[0]/2-((lbda_crop[0]-dlbda_crop[0]/2)%0.2)
max_tick = lbda_crop[-1]+dlbda_crop[-1]/2+(0.2-((lbda_crop[-1]+dlbda_crop[-1]/2)%0.2))
major_ticks1 = np.arange(min_tick,max_tick,maj_tick_sp)
minor_ticks1 = np.arange(min_tick,max_tick,min_tick_sp)
ax1.set_xticks(major_ticks1)
ax1.set_xticks(minor_ticks1, minor = True)
ax1.tick_params(which = 'both', direction = 'in')
ax2.set_xticks(major_ticks1)
ax2.set_xticks(minor_ticks1, minor = True)
ax2.tick_params(which = 'both', direction = 'in')
# Plot measured spectrum
ax1.errorbar(lbda_crop, lbda_crop*spec_crop,
lbda_crop*spec_err_crop, fmt=cols[0]+'o',
label='Measured spectrum')
ax1.legend(loc='best')
ax2.errorbar(lbda_crop, lbda_crop*spec_crop,
lbda_crop*spec_err_crop, fmt=cols[0]+'o',
label='Measured spectrum')
ax2.legend(loc='best')
#plt.savefig("Template_fit.pdf", bbox_inches='tight')
plt.show()
Fits HDU-0 data and header successfully loaded. Data shape: (3, 4142)
Fits HDU-0 data and header successfully loaded. Data shape: (3, 4145)
Fits HDU-0 data and header successfully loaded. Data shape: (3, 4153)